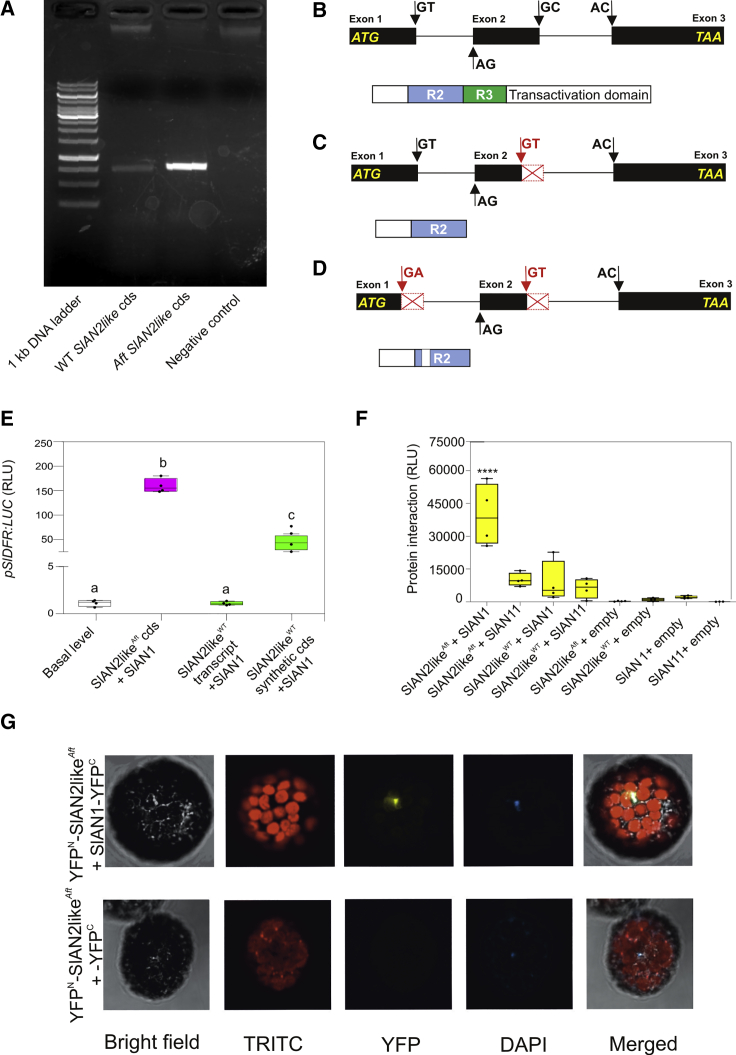
Figure 4.
Structural and Functional Analysis of the SlAN2like Factors Produced from the Transcripts Identified in WT and Aft Fruit Peel.
(A) Agarose gel electrophoresis of the RT–PCR products showing the SlAN2like transcripts amplified from WT and Aft fruit peel cDNAs. The expected length of the WT SlAN2like CDS (Solyc10g086290.1.1) is 798 bp.
(B) Schematic representation of intron–exon structure of the WT genomic sequence of SlAN2like with the positions of the “canonical” splicing sites (black arrows), which produce the theoretical transcript registered in the SOL Genomics Network database (Solyc10g086290.1.1) (above), and protein produced from its mature mRNA with major functional domains (below). Gene and protein sequences are shown at different scales.
(C) Schematic representation of intron–exon structure of the WT genomic sequence of SlAN2like with the positions of the “canonical” splicing sites (black arrows) and the alternative ones (red arrows), which produce the first shorter transcript identified in fruit peel (above), and protein produced from its mature mRNA with major functional domains (below). Gene and protein sequences are shown at different scales.
(D) Schematic representation of intron–exon structure of the WT genomic sequence of SlAN2like with the positions of the “canonical” splicing sites (black arrows) and the alternative ones (red arrows), which produce the second shorter transcript identified in fruit peel, and protein produced from its mature mRNA with major functional domains (below). Gene and protein sequences are shown at different scales.
(E) Transactivation of the SlDFR promoter driving the firefly luciferase gene in protoplasts with effector plasmids containing the SlAN2like transcripts cloned in WT and Aft fruit peel and the SlAN2like synthetic CDS (corresponding to the theoretical transcript produced from the WT pre-mRNA using the “canonical” splicing sites used in the processing of the pre-mRNA of SlAN2likeAft). MYB proteins were expressed in combination with SlAN1. Data are expressed as relative luciferase activity (RLU) (FireflyLuc/RenillaLuc) with the value of the promoter basal level set to 1 and are means of four biological replicates ± SE. One-way ANOVA with Tukey's HSD post hoc test was performed. Different letters indicate significant differences at P ≤ 0.05.
(F) Split-luciferase complementation assay in WT protoplasts expressing the fusion proteins NLuc-SlAN2likeWT or NLuc-SlAN2likeAft with CLuc-SlAN1 or CLuc-SlAN11. Combinations of each construct with the empty vectors expressing the complementary half of the luciferase gene represent negative controls. Data are expressed as RLU and are means of four biological replicates ± SE. One-way ANOVA with Tukey's HSD post hoc test was performed. Each box was compared with the first one, with significant difference at ****P ≤ 0.0001.
(G) Bimolecular fluorescence complementation assay analyzing the interaction between SlAN2likeAft and SlAN1 in tomato protoplasts expressing the fusion proteins YFPN-SlN2likeAft and YFPC-SlAN1. As a control, YFPC-half protein was expressed in combination with YFPN-SlAN2likeAft fusion protein.