Figure 4.
Different Substructures and Distributions of XA1, XA1-2, XA14, and RGAF.
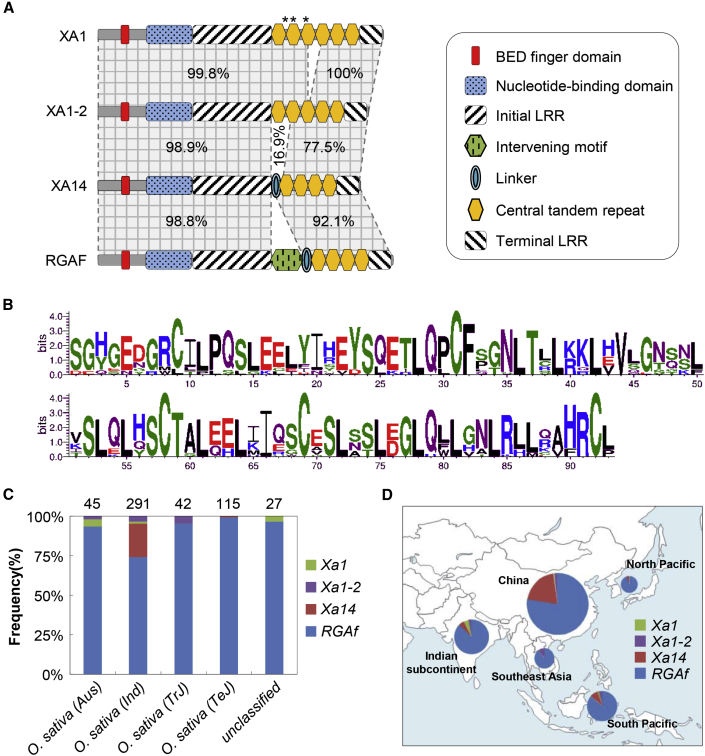
(A) Domains and motifs in XA1, XA1-2, XA14, and RGAF. Percentages indicate sequence identity. Stars indicate missense mutations.
(B) Conserved amino acid residues in the CTRs of the four proteins shown by WebLogos.
(C) Ratios of Xa1, Xa1-2, Xa14, and RGAf alleles in the rice variety collection. Numbers above the columns indicate the total accession numbers of the corresponding subpopulation.
(D) Geographic distribution of Xa1, Xa1-2, Xa14, and RGAf haplotypes in tropical and temperate regions in Asia. Relative frequencies of the haplotypes in each area are represented by the colored sectors of the pie chart. The size of the pie chart is proportional to the number of varieties.