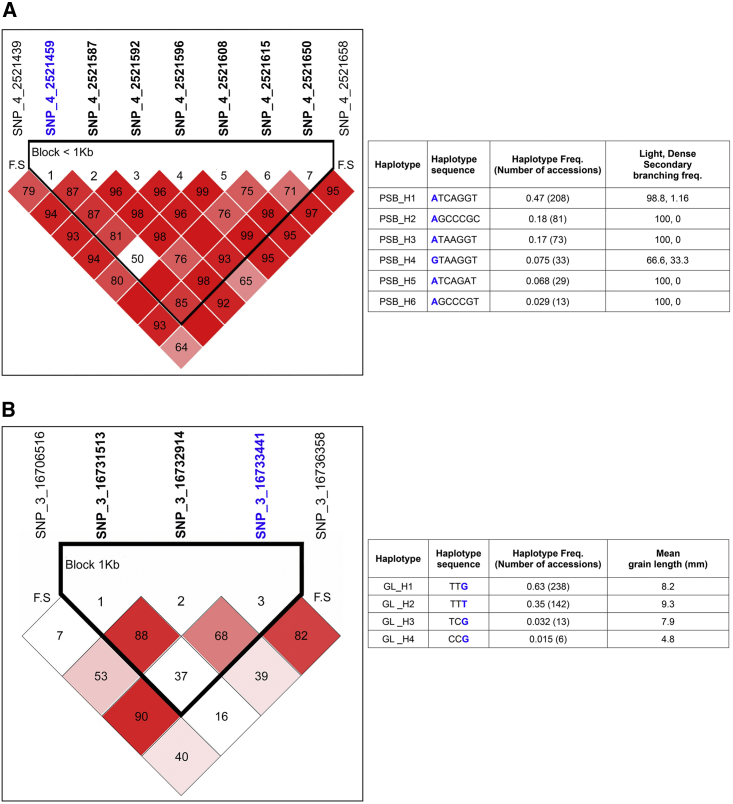
Figure 7.
Linkage Disequilibrium and Haplotype Analysis.
(A) Depiction of strong linkage disequilibrium (LD) on chromosome 4 and the haplotype block containing the GWAS-identified SNP for panicle secondary branching (PSB). The table shows the distribution of various haplotypes for the PSB trait in the mini-core CC3 population.
(B) Depiction of strong LD on chromosome 3 and the haplotype block containing the GWAS-identified SNP for grain length (GL). The table shows the distribution of various haplotypes for the GL trait in the mini-core CC3 population. The GWAS-identified SNP ID is highlighted in blue. The SNP ID in black bold format depicts the block comprising SNP. F. S denotes the block flanking SNP. Red blocks, D′ (normalized LD measure or D) ≤1.0, with logarithm of odds (LOD) score ≥2.0; white blocks, D′ < 1.0 with LOD < 2.0; blue blocks, D′ = 1.0 with LOD < 2.0. Numbers in blocks denote D′ values. The genomic organization is described above the LD plot. LOD was defined as log10(L1/L0), where L1 = likelihood of the data under LD, and L0 = likelihood of the data under linkage equilibrium.