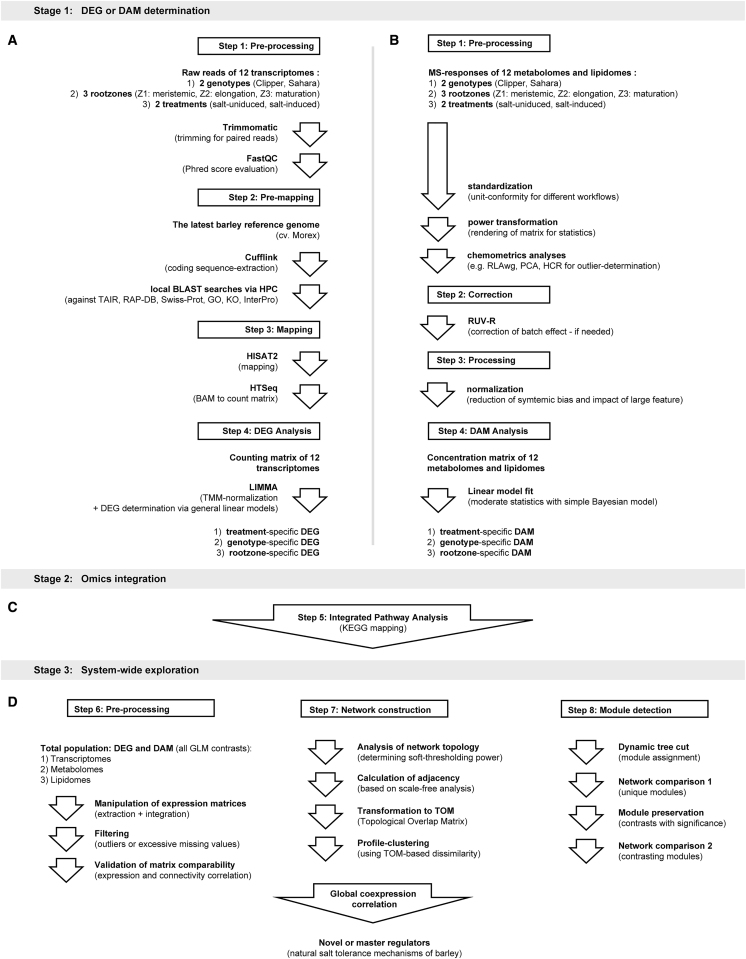
Figure 1.
Overview of the Bioinformatics Pipelines Implemented in This Study.
(A) Pre-processing, pre-mapping, mapping, and DEG analysis of 12 transcriptomes.
(B) Pre-processing, data-correction, normalization, and DAM analysis of 12 metabolomes and lipidomes.
(C) KEGG-based integrated pathway analysis of transcriptomes, metabolomes, and lipidomes.
(D) Pre-processing, network construction, and module detection for global co-expression correlation analysis of multi-omes in barley.
DAM, differentially abundant metabolite; DEG, differentially expressed genes; GLM, general linear model; GO, Gene Ontology; HCR, hierarchical clustering; HPC, high performance computation; KO, Kyoto Encyclopedia of Genes and Genomes Ontology; MS, Mass Spectrometry; PCA, principal component analysis; RAP-DB, Rice Annotation Project - Database; RLAwg, within-group relative log adjustment; TAIR, The Arabidopsis Information Resource; TMM, trimmed mean normalization; Z1, zone 1 (meristematic zone); Z2, zone 2 (elongation zone); Z3, zone 3 (maturation zone).