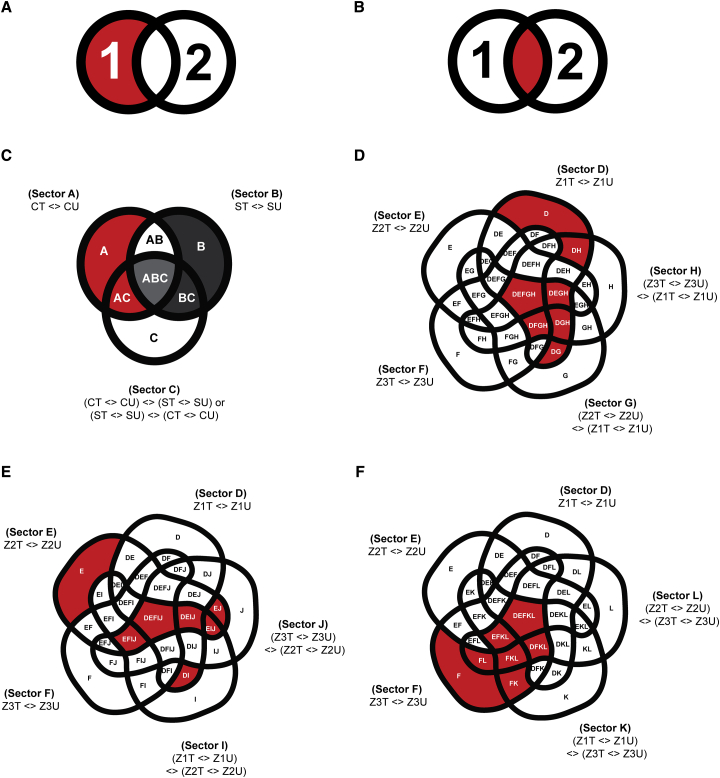
Figure 2.
Design of GLM and Subsetting for the DEG or DAM Determination.
(A and B) Two possible sets of DEGs or DAMs that could account for the genotype- or rootzone-specific phenotypes (colored in red): (A) DEGs or DAMs unique to one genotype or rootzone; (B) DEGs or DAMs common to both genotypes or rootzones, but showed significant contrast in expression or abundance between the two. Numbers 1 and 2 in the figures denote two sets of DEGs or DAMs from two different genotypes/rootzones in comparison.
(C) Genotype-specific DEGs or DAMs for each rootzone. Subsectors correspond to the Clipper-specific DEGs/DAMs (including subsectors A, AC) and Sahara-specific DEGs or DAMs (including subsectors B, BC) in each rootzone are highlighted in red and dark gray, respectively. Subsector ABC are common to both Clipper and Sahara (colored in light gray), but defined by GLM contrast in opposite directions: (CT <> CU) <> (ST <> SU), and (ST <> SU) <> (CT <> CU), respectively.
(D–F) Rootzone-specific DEGs or DAMs of Clipper/Sahara at (D) meristematic zone (Z1), (E) elongation zone (Z2), and (F) maturation zone (Z3), respectively, and with the corresponding subsectors highlighted in red. Symbol “< >” denotes a “contrast/comparison” being tested during differential analysis through fitting of GLM.
CT, salt-treated Clipper; CU, untreated Clipper; DEGs, differentially expressed genes; DAMs, differentially abundant metabolites; ST, salt-treated Sahara; SU, untreated Sahara; Z1T, salt-treated Z1; Z1U, untreated Z1; Z2T, salt-treated Z2; Z2U, untreated Z2; Z3T, salt-treated Z3; Z3U, untreated Z3; <>, contrast of GLM.