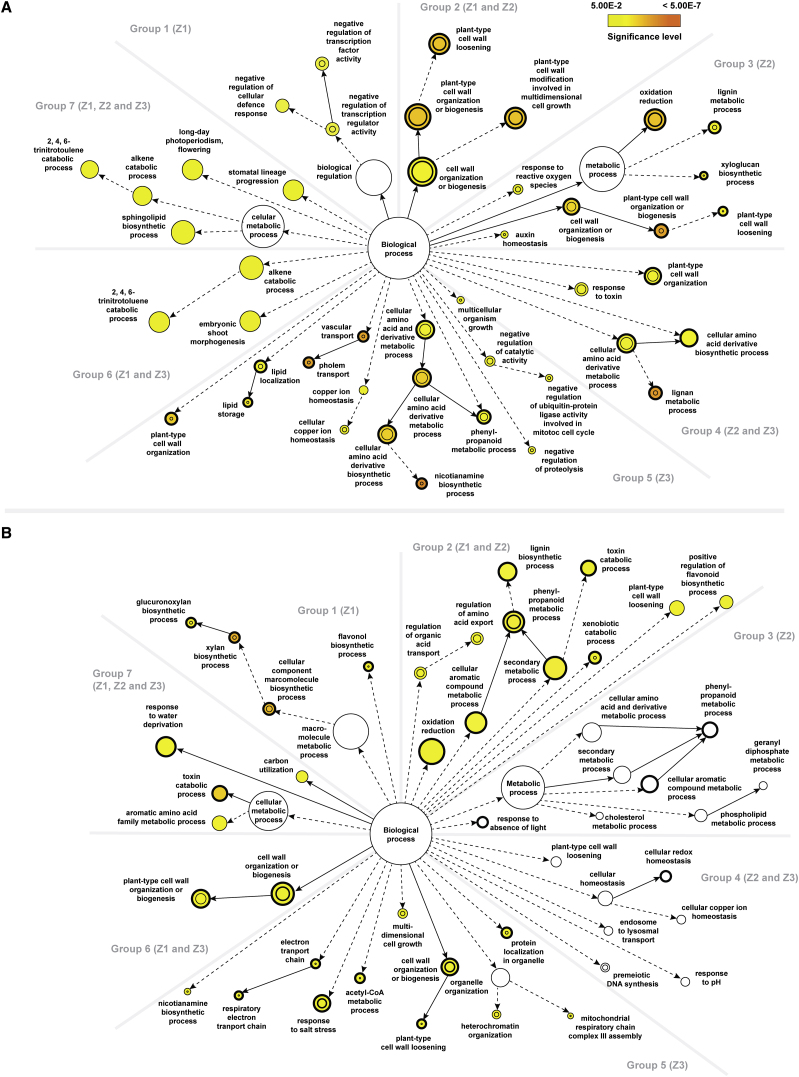
Figure 3.
Comparisons of the Statistical Over-Representation of GO Categories between Different Root Zones of the Two Barley Genotypes upon Salt Stress.
(A) Statistically over-represented GO categories unique to or shared between different rootzones of Clipper.
(B) Statistically over-represented GO categories unique to or shared between different rootzones of Sahara.
Group 1 composed of DEGs found only in Z1. GO analysis via BiNGO (Maere et al., 2005) and REVIGO (Supek et al., 2011) revealed that the most significant over-representation of this group were regulation of transcription and cellular defense response genes in Clipper, and biosynthesis of hemicelluloses, including xylan and its derivatives in Sahara. Group 2 included DEGs found in both Z1 and Z2. GO analysis indicated the genes to be mostly enriched in cell wall modification (in particular cell wall loosening) for Clipper, and phenylpropanoid metabolism for Sahara. Group 3 consisted of DEGs found only in Z2. While plant-type cell wall organization as well as lignin metabolism genes were strongly over-represented in Clipper, no significant enrichment of any GO category could be detected in Sahara. Group 4 represented DEGs found in Z2 and Z3. Lignan metabolism genes and related processes were drastically enriched in Clipper, but similar to Group 3, no significant over-representation was detected in Sahara. Group 5 consists of DEGs found only in Z3. In Clipper, nicotianamine metabolic process as well as vascular transport genes were ranked top in the overrepresentation list, compared with the enrichment of genes encoding proteins targeted to the mitochondrion, response to salt stress, and cell wall organization (xyloglucan metabolism) in Sahara. Group 6 represented DEGs found in both Z1 and Z3. This cluster was enriched in trinitrotoluene catabolism and related processes for Clipper, cell wall organization, or biosynthesis for Sahara. Group 7 contains DEGs found in all three rootzones. Sphingolipid biosynthesis genes were enriched in Clipper, whereas toxin metabolism was the most significantly overrepresented GO category in Sahara. Nodes represent GO categories and node-size is proportional to the number of detected genes for each node. Categories under the same GO hierarchy are linked by interconnected arrows (known as edges) and intensity of node-color indicates the significance level of statistical overrepresentation determined by Fisher's exact test with adjusted p < 0.05 as cutoff as per legend. For reference only, a threshold of 0.2 is set for those sectors showing no significant over-representation and white-colored nodes are used to visualize those ontologies closed to the threshold. Dotted edges indicate one or more hierarchies of GO, which have no statistical significance in the over-representation test and are determined as redundant via REVIGO, were not shown for clarity. DEGs statistically over-represented in both treatment-specific and genotype-specific analyses are denoted by nodes with thickened outlines. DEGs statistically over-represented in both treatment-specific and rootzone-specific batches are denoted by inner circle of nodes. Z1, zone 1 (meristemic zone); Z2, zone 2 (elongation zone); Z3, zone 3 (maturation zone).