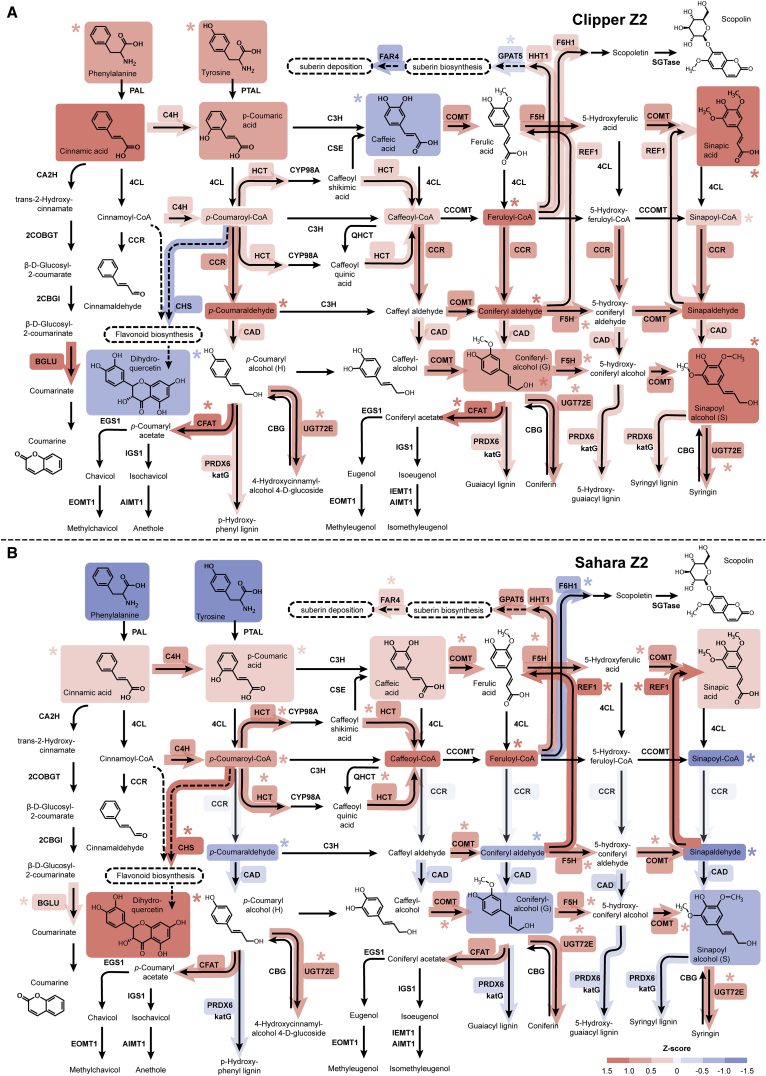
Figure 5.
Standardized Abundance of Transcripts and Metabolites Involved in Phenylpropanoid Biosynthesis at the Elongation Zone (Z2) of the Two Barley Genotypes under Salt Stress.
(A) The abundance of transcripts and metabolites involved in the biosynthesis at Clipper Z2.
(B) The abundance of transcripts and metabolites involved in the biosynthesis at Sahara Z2.
Standardized abundances of transcripts and metabolites shown are the Z scores for TMM-normalized CPM and median-normalized concentration respectively. Level of the standardized abundance (i.e., positive, negative, and zero Z score) is indicated by intensity of shading in red, blue, and pale gray, respectively. Asterisks denote statistically significant differentiation of transcript- and metabolite-abundance (with Benjamini–Hochberg adjusted p < 0.05) after the salt stress compared with the untreated control. Standardized abundance of only the transcripts with significant degree of sequence similarities to the characterized homologs (E < 1.00E-3) and the metabolites within the limit of detection of methodologies and instrumentations adopted in this study are shown. Abundance details of these pathway components at different rootzones of the two barley genotypes before and after the salt treatment can be found in Supplemental Figures 5 and 6.
AIMT1, trans-anol O-methyltransferase; BGLU, beta-glucosidase; CA2H, cinnamic acid 2-hydroxylase; CBG, coniferin beta-glucosidase; 2CBGI, 2-coumarate β-D-glucoside isomerase; CAD, cinnamyl-alcohol dehydrogenase; CCOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CFAT, coniferyl alcohol acyltransferase; C3H, p-coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CHS, chalcone synthase; 4CL, 4-coumarate-CoA ligase; COBGT, 2-coumarate O-beta-glucosyltransferase; COMT, caffeic acid 3-O-methyltransferase; CSE, caffeoylshikimate esterase; CYP98A, coumaroylquinate(coumaroylshikimate) 3′-monooxygenase; EGS1, eugenol synthase; EOMT1, eugenol/chavicol O-methyltransferase; F5H, ferulate-5-hydroxylase; F6H1, feruloyl-CoA ortho-hydroxylase; FAR4, fatty acid reductase 4; GPAT5, glycerol-3-phosphate acyltransferase 5; HCT, shikimate O-hydroxycinnamoyltransferase; HHT1, hydroxyacid O-hydroxycinnamoyltransferase 1; IEMT1, (iso)eugenol O-methyltransferase; IGS1, isoeugenol synthase; katG, catalase-peroxidase; PAL, phenylalanine ammonia-lyase; PTAL, phenylalanine/tyrosine ammonia-lyase; PRDX6, peroxiredoxin 6; REF1, coniferyl-aldehyde dehydrogenase; SGTase, scopoletin glucosyltransferase; QHCT, quinate O-hydroxycinnamoyltransferase; UGT72E, coniferyl alcohol glucosyltransferase.