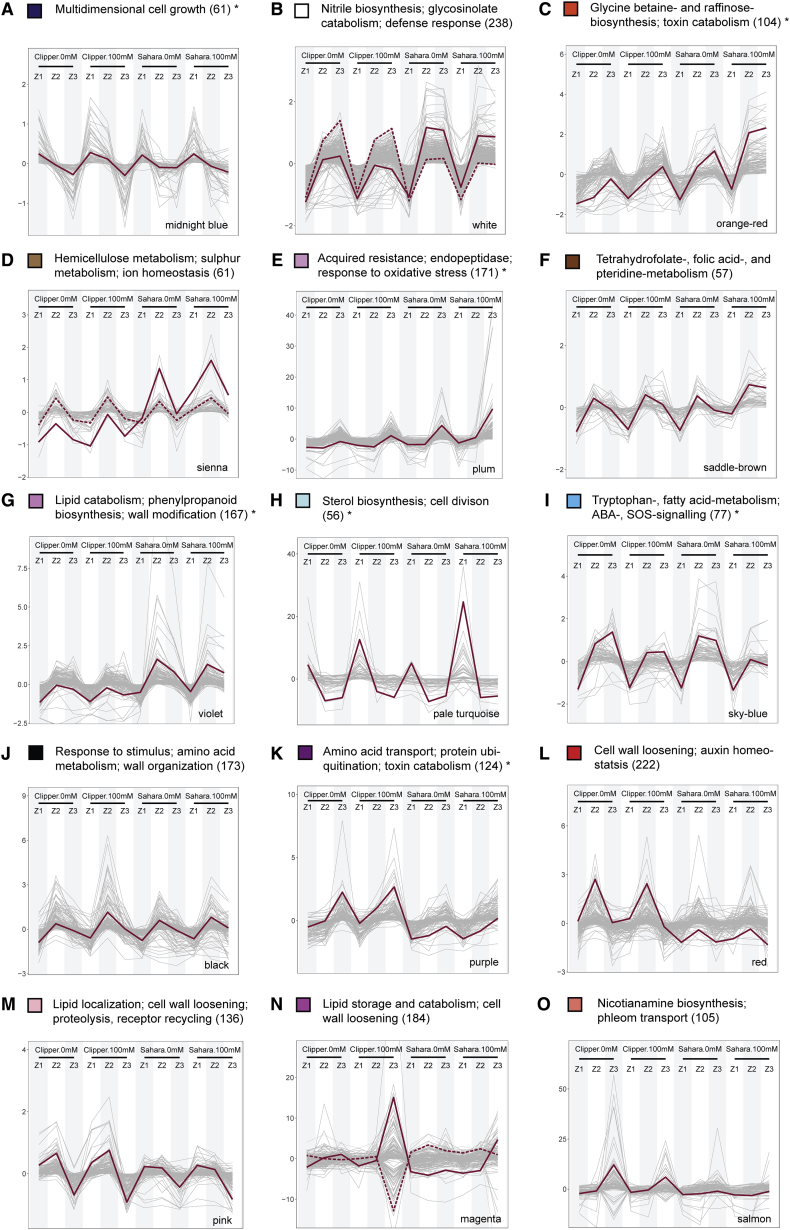
Figure 6.
Selected Modules of Weighted Coexpression Correlation Networks Showing Abundance Profiles of Transcripts and Metabolites.
(A) The abundance profile unique to Clipper.
(B–I) The abundance profiles unique to Sahara.
(J–O) The abundance profiles significantly contrast between the two barley genotypes.
Profiles showing either positive or negative correlations by clustering abundance into differently colored modules through weighted correlation networks. Additional profiles with less obvious differentiation between the two genotypes can be found in Supplemental Figure 11. The color of each module is consistent with Supplemental Figure 10. The most representative trend or centroid of each module represented by solid lines are determined by k-mean clustering (distance method: Pearson) with optimal number of clusters calculated from within-group sum of square method (Madsen and Browning, 2009). Second, the most representative centroid (if any) is indicated by a dotted line. Only expression profiles within 99th percentile are shown for clarity. Annotation of each co-expression clusters are determined by means of the statistical enrichment of GO categories below the cutoff (adjusted p ≤ 0.05) and specific biological role of each module specified here is designated by manual curation of the enrichment outcomes. Asterisks denote the clusters with no significant over-representation and annotations assigned to these clusters are the GO categories with the highest possible level of significance (Supplemental Data 8). Annotated lists of members for each module with significant match (E < 1.00E-4) against TAIR10 genome release (version: June 2016) ranked in descending order according to kME of members can be found in Supplemental Data 9.