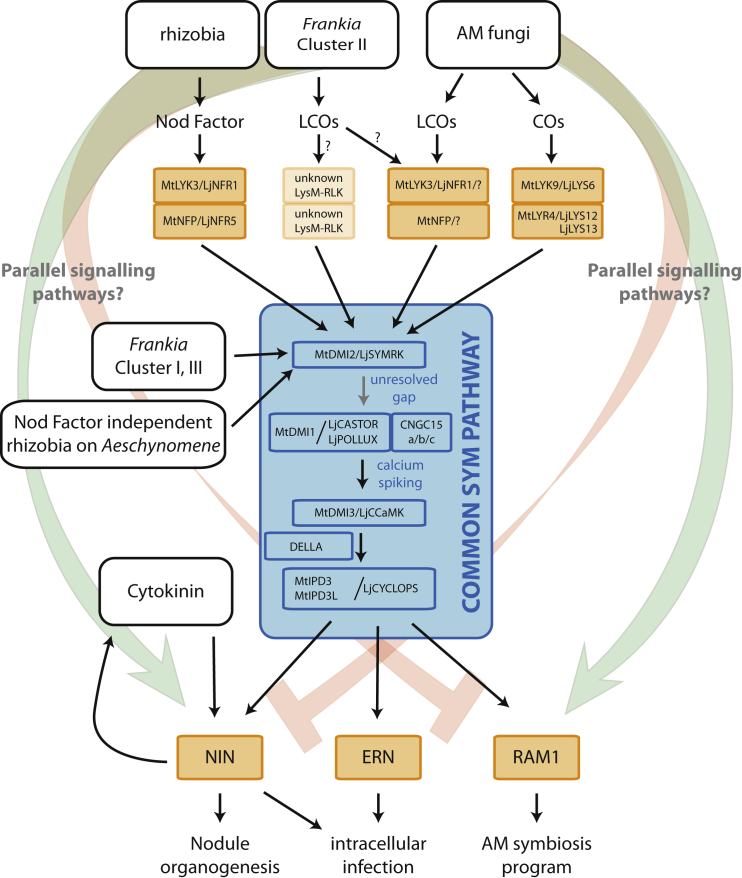
Figure 3.
Schematic Representation of the Common SYM Pathway.
Lipochitooligosaccharides (LCOs) of rhizobia (Nod factors), Frankia cluster II, and arbuscular mycorrhizal (AM) fungi (Myc factors), as well as chitin oligomers (COs) of AM fungi, are perceived by different heterodimeric complexes of LysM-RLKs. These complexes interact also with the leucine-rich repeat-type SYMBIOSIS RECEPTOR KINASE (LjSYMRK/MtDMI2), thereby activating the common SYM pathway. In the case of LCO-independent Frankia clusters I and III as well as Aeschynomene legumes, SYMRK is activated by an as-yet unknown mechanism. Nuclear Ca2+ oscillations are a hallmark of symbiotic signaling induced by rhizobia, Frankia, and AM fungi. Ca2+ oscillations are decoded by calcium-/calmodulin-dependent kinase (CCaMK), leading to activation of MtIPD3/LjCYCLOPS. Downstream of this transcription factor a symbiosis-specific transcriptional network is activated, which includes ERN1 and the NIN–cytokinin feed-forward loop, in the case of nodule formation in legumes, and RAM1 in the case of AM symbiosis. To allow such symbiosis-specific transcriptional activation, parallel pathways are predicted, which can include activating (green) or inhibiting (red) activity.