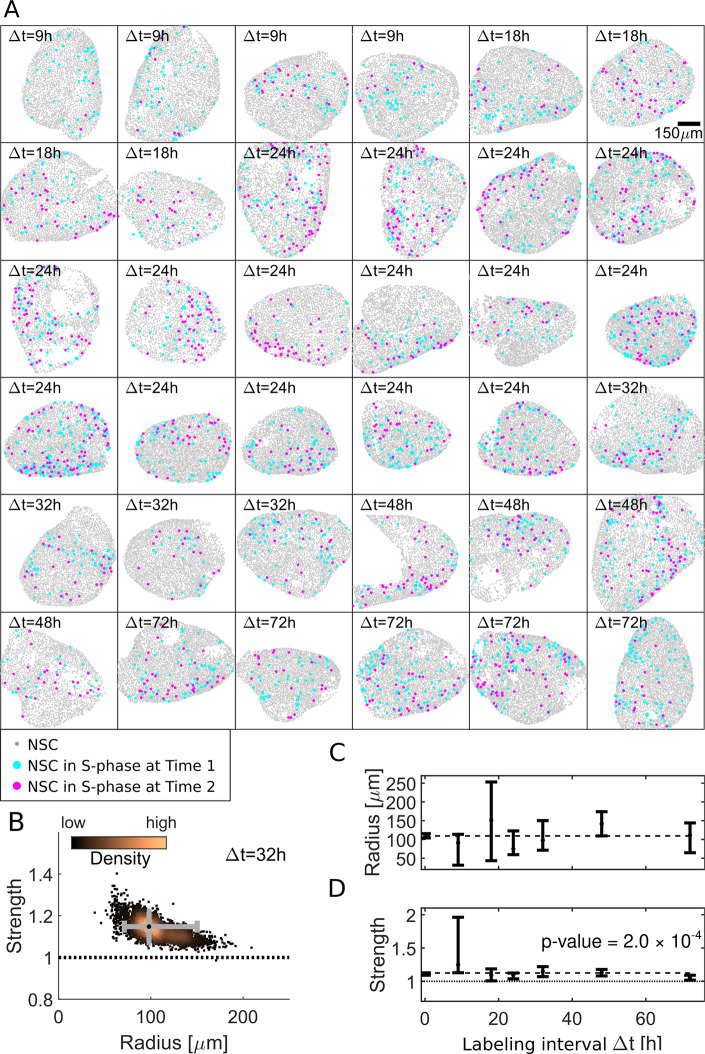
Fig 2. Computational approach identifies an approximately 100-μm aggregation radius of NSCs in S-phase.
(A) Our dataset comprises 36 hemispheres with labeling intervals from Δt = 9 h to 72 h. (B) Posterior sampling identifies the most likely interaction strength of 1.15 and most likely interaction radius of 98 μm for 4 Δt = 32 h hemispheres. Whiskers (gray) cover the 95% CIs for strength and radius. Sampling point density is visualized from copper (high) to black (low). (C) Applied to all 36 hemispheres posterior sampling reveals an interaction radius around 100 μm. (D) The interaction strength is significantly above 1 for all labeling intervals Δt (p-value = 0.0002 for constant fit to most likely values) thus inducing aggregated patterns. CI, confidence interval; NSC, neural stem cell.