SUMMARY
Background:
Candida auris, a multi-drug-resistant fungal pathogen, has become an emerging threat in healthcare settings around the world. Reliable disinfection protocols specifically designed to inactivate C. auris are essential, as many chemical disinfectants commonly used in healthcare settings have been shown to have variable efficacy at inactivating C. auris.
Aim:
Ultraviolet germicidal irradiation (UVGI) was investigated as a method to inactivate clinically relevant strains of C. auris.
Methods:
Ten C. auris and two C. albicans isolates were exposed to ultraviolet (UV) energy to determine the UV dose required to inactivate each isolate. Using a UV reactor, each isolate (106 cells/mL) was exposed to 11 UV doses ranging from 10 to 150 mJ/cm2 and then cultured to assess cell viability.
Findings:
An exponential decay model was applied to each dose–response curve to determine inactivation rate constants for each isolate, which ranged from 0.108 to 0.176 cm2/mJ for C. auris and from 0.239 to 0.292 cm2/mJ for C. albicans. As the model of exponential decay did not accurately estimate the dose beyond 99.9% inactivation, a logistic regression model was applied to better estimate the doses required for 99.999% inactivation. Using this model, significantly greater UV energy was required to inactivate C. auris (103–192 mJ/cm2) compared with C. albicans (78–80 mJ/cm2).
Conclusion:
UVGI is a feasible approach for inactivating C. auris, although variable susceptibility among isolates must be taken into account. This dose–response data is critical for recommending UVGI dosing strategies to be tested in healthcare settings.
Keywords: Candida auris, Emerging pathogen, Drug resistance, Ultraviolet germicidal irradiation, Disinfection, Ultraviolet inactivation
Introduction
Candida auris is a multi-drug-resistant opportunistic yeast pathogen that was characterized as a serious global health threat by the Centers for Disease Control and Prevention (CDC) in 2016. C. auris has been identified in over 30 countries, and has recently emerged as a threat in the US healthcare system, where 950 clinical cases had been confirmed in 14 states as of 30th November 2019 [1]. The majority of these cases were associated with healthcare and long-term care facilities in large metropolitan areas in the states of Illinois, New Jersey and New York. As a result, some states have initiated targeted patient screening to control the spread of the organism throughout healthcare environments [1]. These screens have identified an additional 1908 patients in the USA who are colonized but not infected with C. auris. Severe C. auris infections are continuing to occur globally, with CDC estimating a significant mortality rate ranging from 30% to 60% as many infected patients are immunocompromised or have existing medical conditions [2].
C. auris was originally described in 2009 after being isolated from the ear canal of a patient in a Japanese hospital [3]. Identified isolates from three continents have been placed in phylogenetic clades representing distinct geographic regions: South Asia, South Africa, South America and East Asia [4]. Recently, a new isolate was identified in Iran that was phylogenetically separated from the four existing clades, suggesting a potential fifth clade derived from a new geographic region [5]. As these outbreaks continue to occur globally, the complex nature of this organism continues to unfold, highlighting the importance of developing effective infection prevention and control approaches.
Environmental assessment studies conducted within healthcare facilities with active C. auris cases have shown that the yeast can be recovered from a broad variety of environmental surfaces [6,7], and may retain metabolic activity for up to 1 month on hard, non-porous surfaces [8]. Recent studies have shown that chemical disinfectant strategies, particularly those using quaternary ammonium disinfectants, have variable effectiveness [9,10]; however, the Environmental Protection Agency (EPA) has made recent strides in providing guidance on the most effective agents against C. auris. While the current EPA recommendations call for the use of Clostridioides difficile disinfection protocols [11], several disinfectants have received EPA-registered label claims for C. auris. CDC has also been working to determine the most effective agents for use in public health emergencies, and have received an EPA-approved Federal Insecticide, Fungicide, and Rodenticide Act (FIFRA) Section 18 exemption for emergency-case use of seven products effective against C. auris. Exposure of healthcare workers (HCWs) to chemical disinfectants, such as quaternary ammonium compounds, peracetic acid, and combinations of disinfectants such as peracetic acid and hydrogen peroxide, has been shown to cause respiratory symptoms and occupational asthma [12–15]. In addition, personal protective equipment and other safety measures are not always readily available or properly used to protect workers against the harmful effects of these agents. It is important that the health of patients and HCWs is considered when advising on infection control practices. The products approved under CDC’s Section 18 exemption provide alternatives to many of these harmful agents; however, other strategies, such as ultraviolet germicidal irradiation (UVGI), would further limit exposure of HCWs to chemical agents.
Emerging disinfectant technologies based on UVGI are currently being used in conjunction with chemical disinfectant approaches for terminal disinfection within the US healthcare sector, and may offer improved alternative strategies to inactivate C. auris. UVGI devices commonly use low-pressure mercury vapour lamps to emit UV energy in the UV-C band (wavelengths of 100–280 nm), predominantly at 254 nm. Studies conducted by Cadnum et al. demonstrated that, similar to C. difficile, increased exposure times to a UV-C room decontamination device were necessary to achieve 5-log reductions in C. auris viability compared with meticillin-resistant Staphylococcus aureus, a drug-resistant bacterial pathogen that causes healthcare-associated infections [16]. These findings were supported in recent studies assessing other UV-C decontamination devices [17,18]. While these initial studies have demonstrated the feasibility of UVGI approaches for inactivating C. auris, it is critical to determine the UV dose–response relationships to improve guidance on the most effective intervention strategies. The objective of this study was to determine the UV dosages required to inactivate multiple strains of C. auris shown to exhibit varying levels of drug resistance.
Methods
Culture methodology and UV exposure
Ten isolates of C. auris used in this study were obtained from the CDC and FDA Antibiotic Resistance Bank (AR Bank #0381 to #0390) [19]. The isolates represented the four major phylogenetic clades described to date, and varied in their susceptibility to antifungal drugs (Table I). In addition, two Candida albicans strains were obtained from the American Type Culture Collection for testing (ATCC #10231 and ATCC #18804). Candida isolates were cultured on Sabouraud dextrose agar (SDA) at 30 °C for 7–10 days. The isolates were harvested in sterile double distilled de-ionized water, and yeast cells were quantified using a haemocytometer. A suspension of 106 Candida cells per mL was prepared in water, and 3-mL aliquots were exposed to increasing doses of UV-C energy (254 nm) using a dual-collimation aqueous UV reactor as described previously [20]. The doses used were as follows: 10, 20, 30, 40, 50, 60, 70, 80, 90, 100 and 150 mJ/cm2 or μW/cm2. The sample cuvette housing and a radiometer detector were located at opposite ends of the reactor, 27.9 cm from the centrally located mercury lamp. Collimating apertures directed the UV-C beam at the cuvette and detector, which monitored the UV dose applied to each sample. The Candida suspensions were placed in quartz sample cuvettes (1.4 cm2, 0.13-cm wall thickness) that were housed on a magnetic stirrer. Gentle stirring ensured that the Candida cells remained in suspension throughout the UV-C exposure. Following exposure, the suspensions (100 μL) were spiral plated in triplicate on SDA plates using a Spiral Biotech Autoplate Model 3000 plating system (Bethesda, MD, USA). Plates were incubated at 30 °C for 72 h, and the concentration of viable cells remaining in the suspension was quantified using a Spiral Biotech Model 530 Color Q Count. An unexposed suspension served as a control. Three independent UVGI dose–response experiments were performed for each isolate tested.
Table I.
Summary characteristics of Candida isolates used in the study
| Strain | Clade | Clade origin | Antifungal drug susceptibilitya | ||
|---|---|---|---|---|---|
| FLC | CAS | AMB | |||
| C. auris AR Bank #0381b | II | East Asia | S | S | S |
| C. auris AR Bank #0382 | I | South Asia | S | S | S |
| C. auris AR Bank #0383 | III | Africa | R | S | S |
| C. auris AR Bank #0384 | III | Africa | R | R | S |
| C. auris AR Bank #0385 | IV | South America | R | S | S |
| C. auris AR Bank #0386 | IV | South America | R | S | S |
| C. auris AR Bank #0387 | I | South Asia | S | S | S |
| C. auris AR Bank #0388 | I | South Asia | R | S | S |
| C. auris AR Bank #0389 | I | South Asia | R | S | R |
| C. auris AR Bank #0390 | I | South Asia | R | S | R |
| C. albicans ATCC #10231 | - | Unknown | R | S | NA |
| C.albicans ATCC#18804b | - | Uruguay | S | S | NA |
S, sensitive; R, resistant; NA, not available; FLC, fluconazole (triazole class drug); CAS, caspofungin (polyene class drug); AMB, amphotericin B (echinocandin class drug).
Antifungal drug susceptibility was based on tentative breakpoints of the Centers for Disease Control and Prevention [19,39].
Type strains.
Data modelling and statistical analysis
A dose–response curve was plotted using MATLAB R2015a Version 8.5.0.197613 (MathWorks, Inc., Natick, MA, USA) by graphing the survival fraction (S) at each UV dose (D) for each experiment performed. A simple model of exponential decay/inactivation was applied to each dose–response curve to determine the inactivation rate constants (k-values). The k-value is inversely related to the dose required to obtain a specific survival fraction of C. auris. As the k-value increases, the UV dose required to reach a particular reduction in the viability of C. auris decreases:
In addition to the exponential decay model, a logistic regression model was also applied to the dose–response data using JMP Version 13.2.0 (SAS Institute Inc., Cary, NC, USA), where the logit of S was determined as:
The logit of S was then plotted against the natural logarithm of the UV dose. Here, the log odds of survival has a linear predictor comprising b as the y-intercept and a as the regression coefficient for ln(D):
The triplicate k-values as well as the doses required for 99.9% (S=0.001) and 99.999% (S=0.00001) inactivation were calculated for each Candida isolate (N=3/isolate) using the exponential decay model and logistic regression model. The values determined for the 10 C. auris isolates were compared with the two C. albicans strains tested by one-way analysis of variance (ANOVA) followed by Bonferroni’s t-test using Sigma-Plot v. 12.5 (Systat Software, Inc., San Jose, CA, USA). Pairwise comparisons among the C. auris isolates were also conducted using one-way ANOVA followed by Bonferroni’s t-test. P≤0.05 was considered to indicate significance.
Results
Inactivation rate constants were calculated for each of the 10 C. auris isolates exposed to UV-C energy in aqueous solution. The k-values, which are inversely related to the UV dose needed to obtain a specific survival fraction of C. auris, ranged from 0.108 to 0.176 cm2/mJ (Table II). With the exception of AR Bank #0381, all k-values of C. auris were significantly lower than those observed in the C. albicans strains tested (P<0.05). The highest k-value was observed for the C. auris isolate, AR Bank #0381 (Table II). Inactivation rate constants represent the rate at which the organism is inactivated based on the dosage of UV energy applied, and can be used to extrapolate the dosage required to obtain target log reductions (LR) in the viability of C. auris. A 5 LR in viability was observed at predicted UV doses ranging from 66 to 110 mJ/cm2 for the 10 C. auris isolates tested (Table II). The dosages required to inactivate C. auris were higher than those required to inactivate C. albicans (41–49 mJ/cm2). This difference was significant for all strains except for AR Bank #0381 and AR Bank #0388.
Table II.
Model of simple exponential decay: inactivation rate constants (k-values) and predicted ultraviolet germicidal irradiation doses required for the inactivation of Candida auris
| Strain | k-valued (cm2/mJ) | R2 | D99.9d (mJ/cm2) | D99.999d (mJ/cm2) |
|---|---|---|---|---|
| C. auris AR Bank #0381 | 0.176 ± 0.017b | 0.9982 | 39.6 ± 4.0 | 66.0 ± 6.7 |
| C. auris AR Bank #0382 | 0.108 ±0.022a,b,c | 0.9509 | 66.2 ± 15.0a,b,c | 110.3 ± 25.0,b,c |
| C. auris AR Bank #0383 | 0.138 ± 0.010a,b | 0.9476 | 50.1 ± 3.4a,b | 83.5± 5.6a,b |
| C. auris AR Bank #0384 | 0.134 ± 0.019a,b | 0.8430 | 52.5 ± 7.6a,b | 87.4± 12.7a,b |
| C. auris AR Bank #0385 | 0.127 ± 0.001a,b | 0.9093 | 54.3 ± 0.2a,b | 90.6 ± 0.4a,b |
| C. auris AR Bank #0386 | 0.142 ± 0.014a,b | 0.9388 | 49.2 ± 5.0a,b | 81.9 ± 8.3a,b |
| C. auris AR Bank #0387 | 0.144 ± 0.009a,b | 0.9415 | 48.1 ± 2.9a,b | 80.2 ± 4.9a,b |
| C. auris AR Bank #0388 | 0.154 ± 0.025a,b | 0.9271 | 45.8 ± 8.2b | 76.4 ± 13.7b |
| C. auris AR Bank #0389 | 0.130 ± 0.022a,b | 0.9512 | 54.3 ± 9.9a,b | 90.5 ± 16.5a,b |
| C. auris AR Bank #0390 | 0.126 ± 0.015a,b | 0.9939 | 55.5 ± 6.8a,b | 92.5± 11.4a,b |
| C. albicans ATCC #10231 | 0.239 ± 0.023 | 0.8756 | 29.1 ± 2.7 | 48.5 ± 4.5 |
| C. albicons ATCC #18804 | 0.292 ± 0.083 | 0.9987 | 24.9 ± 6.2 | 41.4 ± 10.3 |
D99.9, ultraviolet dose required for 3-log reduction; D99.999, ultraviolet dose required for 5-log reduction.
Significantly different following one-way analysis of variance compared with C. albicans ATCC #10231 (P≤0.034).
Significantly different following one-way analysis of variance compared with C. albicans ATCC #18804 (P≤0.015).
Significantly different following pairwise analysis compared with C. auris AR Bank #0381 (P≤0.014).
The values represent the average of three independent experiments ± standard deviation.
The simple model of exponential decay is useful for calculating UV inactivation rate constants for comparing microorganisms; however, the model did not accurately estimate the UV dose required to obtain a 5 LR in viability for many of the tested C. auris isolates (Figure 1). While the model fit the dose–response of a few isolates fairly well (e.g. AR Bank #0382 shown in Figure 1a), the model underestimated the dose required for 4 and 5 LR for many of the isolates (e.g. AR Bank #0384 shown in Figure 1b). Other models were explored to better extrapolate the UV dose required for 5 LR in viability for the tested C. auris isolates. To overcome the limitations with the exponential decay model, a logistic regression model was applied that better fit the dose–response curves of all isolates (Figure 1a,b). Using this procedure, 3 and 5 LR were predicted for each Candida isolate. The doses calculated for 3 LR using both models were similar, but differed considerably for 5 LR (Tables II and III). As expected based on the model fit, the exponential decay model predicted lower doses than the logistic regression model for 5 LR in viability for all isolates tested (Tables II and III). The predicted doses required for 5 LR in the viability of C. auris remained higher than the doses calculated for C. albicans, but only AR Bank #0382, #0384 and #0385 were significantly different (P≤0.006, Table II). The C. auris isolates tested exhibited variability in their susceptibility to UV energy. The dose required for 5 LR was significantly higher for AR Bank #0385 compared with AR Bank #0386, AR Bank #0387 and AR Bank #0388 (P=0.018, 0.007 and 0.034, respectively).
Figure 1.
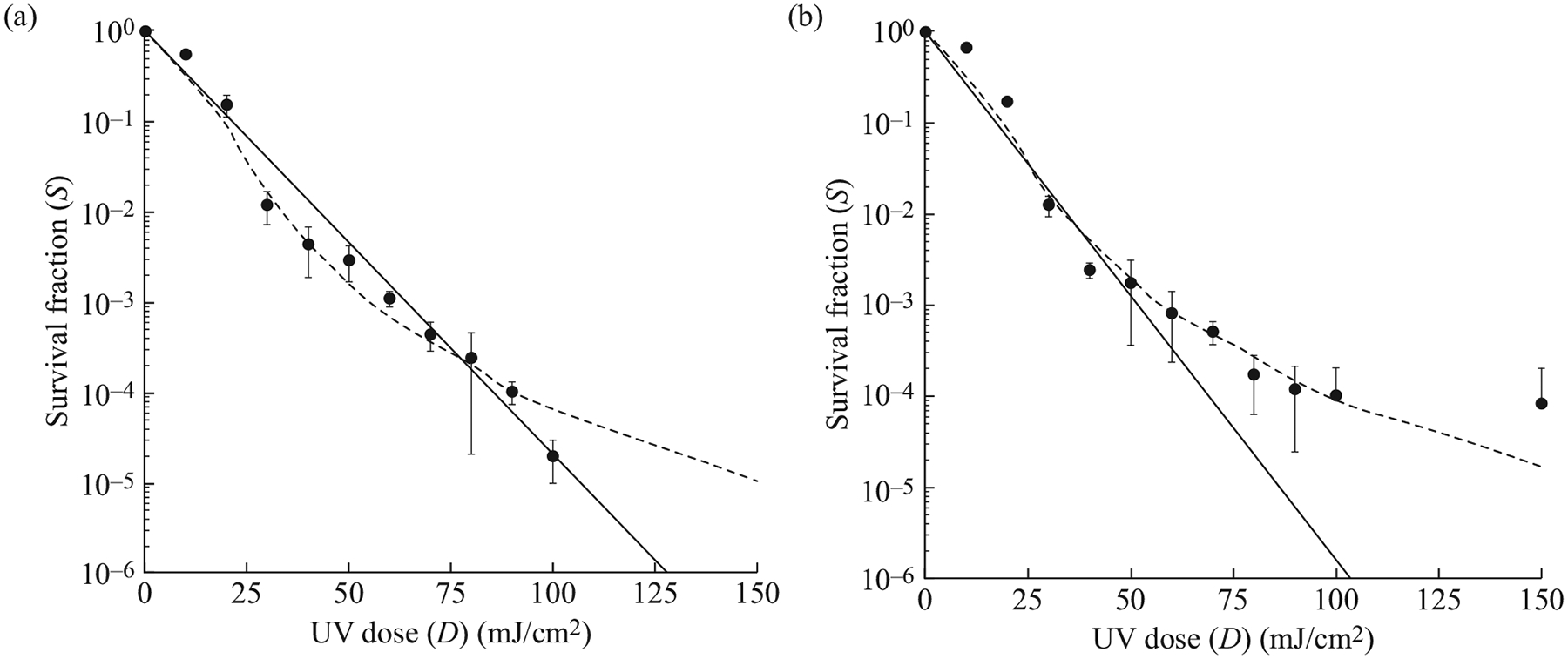
Ultraviolet (UV) dose–response curves for two strains of Candida auris. The survival fraction at each UV dose tested (10–150 mJ/cm2) is represented by black dots for AR Bank #0382 (a) and AR Bank #0384 (b). The error bars represent the standard deviation observed following three independent experiments. The solid lines represent the best-fit line utilizing the exponential decay model. This model fit some strains well (e.g. AR Bank #0382) but underestimated the dose required above 3-log reductions in others (e.g. AR Bank #0384). A logistic regression model, represented by the dashed lines, was applied to the dose–response curves to better estimate the UV dose required for these higher log reductions.
Table III.
Logistic regression: predicted ultraviolet germicidal irradiation doses required for inactivation of Candida auris
| Strain | R2 | D99.9f (mJ/cm2) | D99.999f (mJ/cm2) |
|---|---|---|---|
| C. auris AR Bank #0381 | 0.8555 | 39.0 ± 4.2c,d,e | 121.6 ± 14.5 |
| C. auris AR Bank #0382 | 0.9563 | 54.6 ± 1.4a,b | 148.6 ± 6.2a,b |
| C. auris AR Bank #0383 | 0.9429 | 48.2 ± 4.3a,b | 132.0 ± 20.3 |
| C. auris AR Bank #0384 | 0.9341 | 57.1 ± 6.0a,b | 165.7 ± 38.2a,b |
| C. auris AR Bank #0385 | 0.8625 | 59.4 ± 7.2a,b | 191.6 ± 45.9a,b |
| C. auris AR Bank #0386 | 0.9719 | 43.9 ± 0.5a,b,d,e | 111.1 ± 4.6e |
| C. auris AR Bank #0387 | 0.9575 | 41.1 ± 3.4a,b,c,d,e | 103.1 ± 11.4e |
| C. auris AR Bank #0388 | 0.9406 | 43.0 ± 2.5a,b,d,e | 116.4 ± 18.9e |
| C. auris AR Bank #0389 | 0.9364 | 48.2 ± 0.8a,b | 131.7 ± 11.5 |
| C. auris AR Bank #0390 | 0.9381 | 43.4 ± 4.5a,b,d,e | 126.1 ± 22.7 |
| C. albicans ATCC #10231 | 0.9075 | 30.6 ± 1.7 | 79.6 ± 3.1 |
| C. albicans ATCC #18804 | 0.9142 | 27.1 ± 4.6 | 77.9 ± 4.0 |
D99.9, ultraviolet dose required for 3-log reduction; D99.999, ultraviolet dose required for 5-log reduction.
Significantly different following one-way analysis of variance compared with C. albicans ATCC #10231 (P≤0.037).
Significantly different following one-way analysis of variance compared with C. albicans ATCC #18804 (P≤0.013).
Significantly different following pairwise analysis compared with C. auris AR Bank #0382 (P≤0.028).
Significantly different following pairwise analysis compared with C. auris AR Bank #0384 (P≤0.035).
Significantly different following pairwise analysis compared with C. auris AR Bank #0385 (P≤0.034).
Values represent the average of three independent experiments ± standard deviation.
Discussion
UVGI was employed as a disinfection strategy for inactivating the emerging fungal opportunistic pathogen, C. auris. The panel of isolates obtained from the CDC and FDA Antibiotic Resistance Bank represented all four major phylogenic clades that have been identified in healthcare facilities around the world. Variability in UVGI susceptibility was observed among the 10 C. auris isolates tested; however, the susceptibility of C. auris was much lower than that observed for C. albicans. While the exponential decay model was not the most accurate for UV dose extrapolation, inactivation rate constants are commonly used for UV dose estimation and for comparing UV susceptibility among micro-organisms. The dose–response curves for all Candida strains tested in a water suspension resulted in inactivation rate constants similar to those observed with other Candida species tested in liquid suspension [21–23] as well as Bacillus subtilis spores that were tested using the same National Institute for Occupational Safety and Health UVGI system [20]. Other fungi, such as Aspergillus, Penicillium, Eurotium and Fusarium spp., have been shown to have inactivation rates much lower than C. auris when exposed to UV-C in liquid suspension, which indicates that substantially higher doses are required for inactivation [21,24,25]. This could be due to the difference in pigmentation observed between Candida yeast cells and the dematiaceous amerospores produced by these fungi. The pigmentation of Fusarium oxysporum and Penicillium italicum has been shown to protect the organisms from UV-C, as mutants lacking pigmentation were more susceptible to inactivation following UV-C exposure [25]. This study demonstrated that C. auris was capable of being inactivated by UV in the laboratory at doses similar to or lower than other commonly encountered fungal species. Incorporation of UVGI methods into C. auris terminal disinfection practices could be advantageous. C. auris isolates were previously shown to be inactivated using a UV-C room decontamination device at rates similar to those observed with C. difficile. As such, the existing UVGI cycles utilized for C. difficile disinfection may be sufficient for terminal disinfection of environments contaminated with C. auris, although some UVGI systems may require longer exposure times to reach the lethal dose [16,26].
Interestingly, the rate of inactivation in the present study was highest for AR Bank #0381, resulting in lower UV doses required to inhibit cell growth. This isolate was originally isolated from the ear canal of a patient in Japan [3], and, according to the CDC and FDA Antibiotic Resistance Isolate Bank, is the most susceptible to antifungal drugs compared with the other isolates in the panel. These findings are consistent with recent studies demonstrating the uniqueness of this strain and others belonging to Clade II, which are typically associated with ear infections and not the invasive infections caused by members of the other clades [27]. It was observed that those strains requiring higher minimum inhibitory concentrations of antifungal drugs also required higher UV doses for inactivation (i.e. AR Bank #0384 and AR Bank #0385). This trend is similar to what has been observed with multi-drug-resistant Escherichia coli in wastewater that require higher UV doses to inhibit growth than antibiotic-susceptible E. coli [28].
Seven of the 10 isolates tested exhibited dose–response relationships that were not well represented using a standard exponential decay model. This resulted in the underestimation of UV doses required for 4 and 5 LR in the viability of C. auris. A logistic regression model was applied to the dose–response data that better represented the dose–response relationships of all the isolates tested. Many of the trends observed using the exponential decay model remained true following application of the logistic regression model; however, the ability to accurately extrapolate the doses required for 4 and 5 LR was more consistent using the logistic regression approach. This is an important consideration to take into account in future studies assessing the efficacy of UVGI for fungal opportunistic pathogens. While inactivation rate constants are a commonly used tool for comparing the effectiveness of UV, using the exponential decay model for advising optimal doses for target reductions in microbial viability may not be optimal.
Controlling C. auris outbreaks to date has been challenged by the complex nature of the fungal species. C. auris is often misidentified as other yeast species, such as C. haemulonii, C. sake, C. guilliermondii, C. lusitaniae, C. parapsilosis and Rhodotorula glutinis, using traditional yeast identification methods [29]. More specialized methods, such as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and molecular detection methods, have been developed recently for more sensitive and specific identification of C. auris [30]. In addition, a number of outbreak strains are resistant to one or more commonly administered antifungal drugs [4]. Most of the isolates tested in this study were resistant to at least one antifungal drug, with seven of the 10 isolates being resistant to fluconazole (Table I). The increased resistance of C. auris to antifungal drugs compared with other species that cause candidiasis, such as C. albicans, has made it critical to correctly identify the source organism to species level to administer effective treatment regimens and advise appropriate disinfection strategies [30].
Studies assessing several chemical disinfectant strategies for C. auris have demonstrated variable effectiveness, particularly with the use of quaternary ammonium compounds [9,10]. There also seems to be variability in disinfectant susceptibility between C. auris isolates [9,31]. Chlorine-based disinfectants appear to be consistently effective across studies [10,31,32]; however, the exposure of HCWs to chlorine-releasing agents has been shown to be associated with occupational asthma [33,34]. Exposure of HCWs to other commonly used chemical disinfectants, such as quaternary ammonium compounds, peracetic acid, and disinfectant combinations like peracetic acid and hydrogen peroxide, has also been shown to cause respiratory morbidities and occupational asthma [12–15]. Investigating alternative strategies for C. auris disinfection, such as UVGI, could provide a more reproducible disinfection approach to chemical disinfection. Although the use of UV devices can result in the degradation of certain materials, such as plastics [35], corrosive disinfectants could lead to similar levels of aging and degradation. The release of ozone during UV disinfection is also a concern, but studies suggest that the ozone levels in rooms decontaminated with UV devices is minimal [36]. It is important that UVGI of hospital environments is conducted while the room is not occupied, as exposure of patients and HCWs to UV radiation could result in corneal and skin damage, even leading to advanced aging and skin cancers [37]. UVGI technologies have the potential to successfully inactivate harmful micro-organisms in healthcare environments while limiting exposure of HCWs to harmful chemical agents.
This study demonstrated that UVGI is an effective approach for inactivating C. auris using the laboratory-housed UVGI reactor developed at the National Institute for Occupational Safety and Health. Similar to the variability observed with antifungal drug and chemical disinfectant susceptibility, this study also showed variability in UV susceptibility among C. auris isolates. This variability emphasizes the importance of testing a large panel of C. auris isolates to determine effective doses for both UV and chemical disinfection strategies. While these studies were conducted in aqueous solution, they provide preliminary data for designing future studies more relevant to the healthcare setting. Many challenges remain that limit our understanding of the doses of UV that can be delivered to different locations within a healthcare environment using UV-C devices, as well as feasible methods to measure and monitor the doses delivered to various surfaces [38]. To better mimic healthcare surfaces contaminated with C. auris, studies will be conducted using the same UVGI system to inactivate C. auris on non-porous surfaces, such as stainless steel, in an organic soil load as these additional factors will likely affect the ability of UV decontamination devices to reach and penetrate the organism. Additional studies applying these results to UV systems in environments that mimic patient rooms will allow for more accurate recommendations for dosing strategies within healthcare environments.
In conclusion, variability in UVGI susceptibility was observed among the 10 C. auris isolates tested. In addition, the susceptibility of C. auris was lower than that observed for C. albicans. These results highlight the importance of testing the efficacy of the various disinfection strategies, both chemical and UVGI, on multiple isolates of C. auris. In addition, identifying and characterizing the isolates associated with clinical cases is critical not only for effective treatment, but also for determining the most effective disinfection strategies.
Acknowledgements
This project was supported by the National Occupational Research Agenda. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
Funding sources
None.
Footnotes
Conflict of interest statement
None declared.
References
- [1].Centers for Disease Control and Prevention. Tracking Candida auris. Atlanta, GA: CDC; 2019. Available at: https://www.cdc.gov/fungal/diseases/candidiasis/tracking-c-auris.html [last accessed May 2020]. [Google Scholar]
- [2].Centers for Disease Control and Prevention. General information about Candida auris. Atlanta, GA: CDC; 2019. Available at: https://www.cdc.gov/fungal/candida-auris/candida-auris-qanda.html [last accessed August 2019]. [Google Scholar]
- [3].Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 2009;53:41–4. [DOI] [PubMed] [Google Scholar]
- [4].Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 2017;64:134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis 2019;25:1780–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 2016;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lesho EP, Bronstein MZ, McGann P, Stam J, Kwak Y, Maybank R, et al. Importation, mitigation, and genomic epidemiology of Candida auris at a large teaching hospital. Infect Control Hosp Epidemiol 2018;39:53–7. [DOI] [PubMed] [Google Scholar]
- [8].Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 2017;55:2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kean R, Sherry L, Townsend E, McKloud E, Short B, Akinbobola A, et al. Surface disinfection challenges for Candida auris: an invitro study. J Hosp Infect 2018;98:433–6. [DOI] [PubMed] [Google Scholar]
- [10].Cadnum JL, Shaikh AA, Piedrahita CT, Sankar T, Jencson AL, Larkin EL, et al. Effectiveness of disinfectants against Candida auris and other Candida species. Infect Control Hosp Epidemiol 2017;38:1240–3. [DOI] [PubMed] [Google Scholar]
- [11].Centers for Disease Control and Prevention. Infection prevention and control for Candida auris. Atlanta, GA: CDC; 2018. Available at: https://www.cdc.gov/fungal/candida-auris/c-auris-infection-control.html [last accessed May 2020]. [Google Scholar]
- [12].Dumas O, Donnay C, Heederik DJ, Hery M, Choudat D, Kauffmann F, et al. Occupational exposure to cleaning products and asthma in hospital workers. Occup Environ Med 2012;69:883–9. [DOI] [PubMed] [Google Scholar]
- [13].Casey ML, Hawley B, Edwards N, Cox-Ganser JM, Cummings KJ. Health problems and disinfectant product exposure among staff at a large multispecialty hospital. Am J Infect Control 2017;45:1133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hawley B, Casey M, Virji MA, Cummings KJ, Johnson A, Cox-Ganser J. Respiratory symptoms in hospital cleaning staff exposed to a product containing hydrogen peroxide, peracetic acid, and acetic Acid. Ann Work Expo Health 2017;62:28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Walters GI, Burge PS, Moore VC, Thomas MO, Robertson AS. Occupational asthma caused by peracetic acid-hydrogen peroxide mixture. Occup Med (Lond) 2019;69:294–7. [DOI] [PubMed] [Google Scholar]
- [16].Cadnum JL, Shaikh AA, Piedrahita CT, Jencson AL, Larkin EL, Ghannoum MA, et al. Relative resistance of the emerging fungal pathogen Candida auris and other Candida species to killing by ultraviolet light. Infect Control Hosp Epidemiol 2018;39:94–6. [DOI] [PubMed] [Google Scholar]
- [17].de Groot T, Chowdhary A, Meis JF, Voss A. Killing of Candida auris by UV-C: importance of exposure time and distance. Mycoses 2019;62:408–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Maslo C, du Plooy M, Coetzee J. The efficacy of pulsed-xenon ultraviolet light technology on Candida auris. BMC Infect Dis 2019;19:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Centers for Disease Control and Prevention. CDC & FDA Antibiotic Resistance Isolate Bank. Atlanta, GA: CDC; 2017. Available at: https://wwwn.cdc.gov/arisolatebank [last accessed October 2017]. [Google Scholar]
- [20].Martin SB Jr, Schauer ES, Blum DH, Kremer PA, Bahnfleth WP, Freihaut JD. A new dual-collimation batch reactor for determination of ultraviolet inactivation rate constants for microorganisms in aqueous suspensions. J Photochem Photobiol B 2016;162:674–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kowalski W UV rate constants Ultraviolet germicidal irradiation handbook: UVGI for air and surface disinfection. Berlin: Springer; 2009. p. 73–117. [Google Scholar]
- [22].Dolman PJ, Dobrogowski MJ. Contact lens disinfection by ultraviolet light. Am J Ophthalmol 1989;108:665–9. [DOI] [PubMed] [Google Scholar]
- [23].Severin BF, Suidan MT, Engelbrecht RS. Kinetic modeling of U.V. disinfection of water. Water Res 1983;17:1669–78. [Google Scholar]
- [24].Begum M, Hocking AD, Miskelly D. Inactivation of food spoilage fungi by ultra violet (UVC) irradiation. Int J Food Microbiol 2009;129:74–7. [DOI] [PubMed] [Google Scholar]
- [25].Asthana A, Tuveson RW. Effects of UV and phototoxins on selected fungal pathogens of citrus. Int J Plant Sci 1992;153:442–52. [Google Scholar]
- [26].Masse V, Hartley MJ, Edmond MB, Diekema DJ. Comparing and optimizing ultraviolet germicidal irradiation systems use for patient room terminal disinfection: an exploratory study using radiometry and commercial test cards. Antimicrob Resist Infect Control 2018;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Welsh RM, Sexton DJ, Forsberg K, Vallabhaneni S, Litvintseva A. Insights into the unique nature of the East Asian clade of the emerging pathogenic yeast Candida auris. J Clin Microbiol 2019;57:e00007–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang CM, Xu LM, Wang XC, Zhuang K, Liu QQ. Effects of ultraviolet disinfection on antibiotic-resistant Escherichia coli from wastewater: inactivation, antibiotic resistance profiles and antibiotic resistance genes. J Appl Microbiol 2017;123:295–306. [DOI] [PubMed] [Google Scholar]
- [29].Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, et al. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 2017;55:638–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lockhart SR, Jackson BR, Vallabhaneni S, Ostrosky-Zeichner L, Pappas PG, Chiller T. Thinking beyond the common Candida species: need for species-level identification of candida due to the emergence of multidrug-resistant Candida auris. J Clin Microbiol 2017;55:3324–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Abdolrasouli A, Armstrong-James D, Ryan L, Schelenz S. In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris. Mycoses 2017;60:758–63. [DOI] [PubMed] [Google Scholar]
- [32].Moore G, Schelenz S, Borman AM, Johnson EM, Brown CS. Yeasticidal activity of chemical disinfectants and antiseptics against Candida auris. J Hosp Infect 2017;97:371–5. [DOI] [PubMed] [Google Scholar]
- [33].Medina-Ramon M, Zock JP, Kogevinas M, Sunyer J, Torralba Y, Borrell A, et al. Asthma, chronic bronchitis, and exposure to irritant agents in occupational domestic cleaning: a nested case–control study. Occup Environ Med 2005;62:598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Arif AA, Delclos GL. Association between cleaning-related chemicals and work-related asthma and asthma symptoms among healthcare professionals. Occup Environ Med 2012;69:35–40. [DOI] [PubMed] [Google Scholar]
- [35].Cai L, Wang J, Peng J, Wu Z, Tan X. Observation of the degradation of three types of plastic pellets exposed to UV irradiation in three different environments. Sci Total Environ 2018;628–629:740–7. [DOI] [PubMed] [Google Scholar]
- [36].Boyce JM, Havill NL, Moore BA. Terminal decontamination of patient rooms using an automated mobile UV light unit. Infect Control Hosp Epidemiol 2011;32:737–42. [DOI] [PubMed] [Google Scholar]
- [37].Gorman T, Dropkin J, Kamen J, Nimbalkar S, Zuckerman N, Lowe T, et al. Controlling health hazards to hospital workers. New Solut 2013;23(Suppl):1–167. [DOI] [PubMed] [Google Scholar]
- [38].Boyce JM, Donskey CJ. Understanding ultraviolet light surface decontamination in hospital rooms: a primer. Infect Control Hosp Epidemiol 2019;40:1030–5. [DOI] [PubMed] [Google Scholar]
- [39].Centers for Disease Control and Prevention. Antifungal susceptibility testing and interpretation. Atlanta, GA: CDC; 2019. Available at: https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html [last accessed May 2020]. [Google Scholar]


