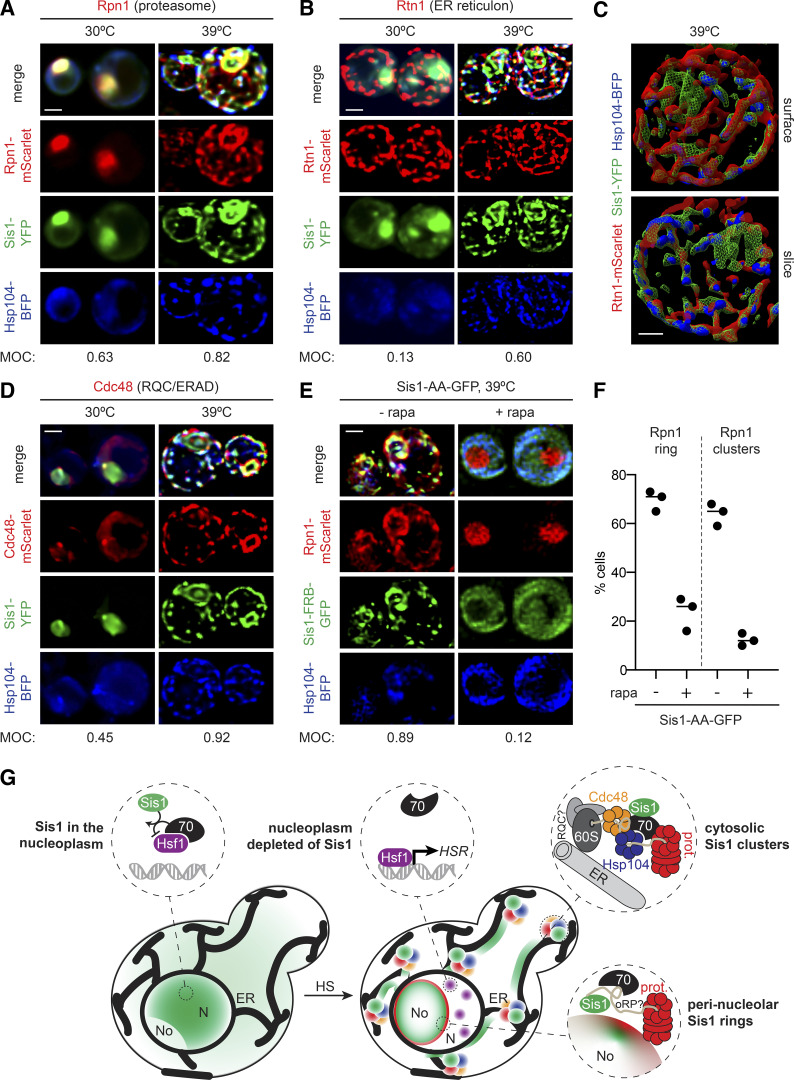
Figure 5.
Sis1 forms an interconnected spatial network with other proteostasis factors during heat shock (HS). (A and B) Deconvolved lattice light-sheet 3D reconstructions of live cells under nonstress conditions and 15 min of HS at 39°C expressing endogenously tagged Sis1-YFP and Hsp104-BFP with Rpn1-mScarlet (A), a subunit of the proteasome; or Rtn1-mScarlet (B), a reticulon protein component of the ER membrane. (C) Top: Space-filling 3D surface rendering of a cell at 39°C showing the association of Sis1-YFP (green) and Hsp104-BFP (blue) with the inside surface of the reticulated ER as marked by Rtn1-mScarlet (red). Bottom: Coronal slice at the midpoint of the cell above. The Sis1-YFP subnuclear ring (green) can be seen and is disconnected from the cytosolic Rtn1 network. Scale bar is 1 µm. (D) Cdc48-mScarlet, an AAA ATPase involved in ERAD and RQC. (E) AA of Sis1 precludes formation the spatial PN. Cells expressing Hsp104-BFP and Rpn1-mScarlet to mark the proteasome in an Sis1-AA background with Sis1-AA-GFP were imaged using a lattice light-sheet microscope following 15 min of HS at 39°C following no pretreatment or pretreatment with rapamycin (rapa) to AA Sis1. In A, B, D, and E, scale bar is 2 µm, and MOC is the fraction of YFP that overlaps with mScarlet. (F) The fraction of cells showing Rpn1 subnuclear rings and cytosolic clusters was quantified during HS in cells with no pretreatment or with rapa pretreatment to AA Sis1. Experiments were performed in triplicate with >20 cells per replicate; the line shows the mean of the replicates. (G) Cartoon model of how the HS-dependent spatial reorganization of the protein homeostasis network is coupled to the regulation of the HSR. Left: In the absence of stress, Sis1 is diffuse throughout the cell and concentrated in the nucleus. In the nucleus, it activates Hsp70 to repress Hsf1. Right: Upon HS, Sis1 relocalizes to the periphery of the nucleolus (No) and the surface of the ER in the cytosol. In the nucleoplasm (N), Hsf1 is now free of Hsp70 and can cluster and activate the HS transcriptional response. Sis1 colocalizes with the proteasome (prot.) at the nucleolar periphery. On the ER, we propose that Sis1 interacts with stalled 60S ribosomes with nascent chains, Cdc48 and possibly the RQC complex, Hsp104, and the proteasome to participate in the resolution of HS-induced cotranslational misfolding.