Figure 1.
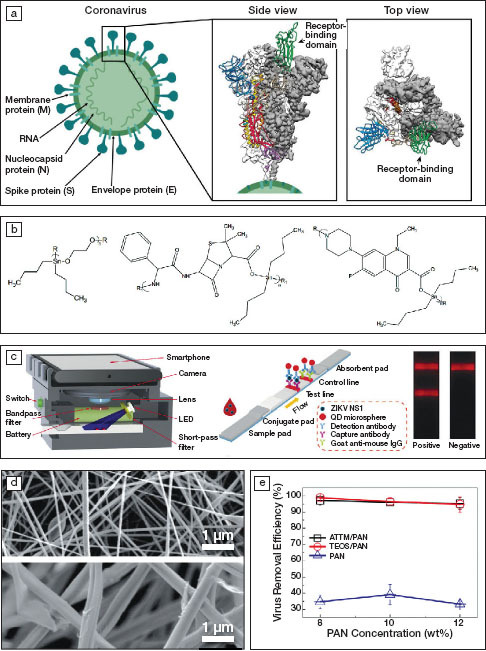
(a) Schematics of a coronavirus and 3D reconstructed views of the structure of its spike protein (S) showing a trimer structure. White or gray areas show the electron density obtained using cryogenic electron microscopy (cryo-EM) and colored areas represent protein conformations, where green corresponds to the receptor-binding domain. Reprinted with permission from References 1 and 2. © 2020 Springer Nature and © 2020 AAAS, respectively. (b) Repeat units of organotin polymers acting as antiviral agents. R and R1 represent side chains. Reprinted with permission from Reference 4. © 2019 Springer Nature. (c) Schematics of a smartphone-based imaging device and quantum-dot (QD)-based fluorescent reader developed for Zika virus detection, with one stripe indicating its presence (test line) and one stripe indicating the test is working (control line using an established antibody; goat anti-mouse IgG). Reprinted with permission from Reference 6. © 2019 Elsevier. (d) Electron micrographs of electrospun membranes containing 8 wt% polyacrylonitrile (PAN, top) and 10 wt% ammonium tetrathiomolybdate (ATTM/PAN, bottom) and (e) the virus removal efficiency as a function of the PAN concentration. TEOS stands for tetraethylorthosilicate. Reprinted with permission from Reference 7. © 2019 Elsevier.
