Figure 2. Mechanisms of Metabolic Control of Cell Fate.
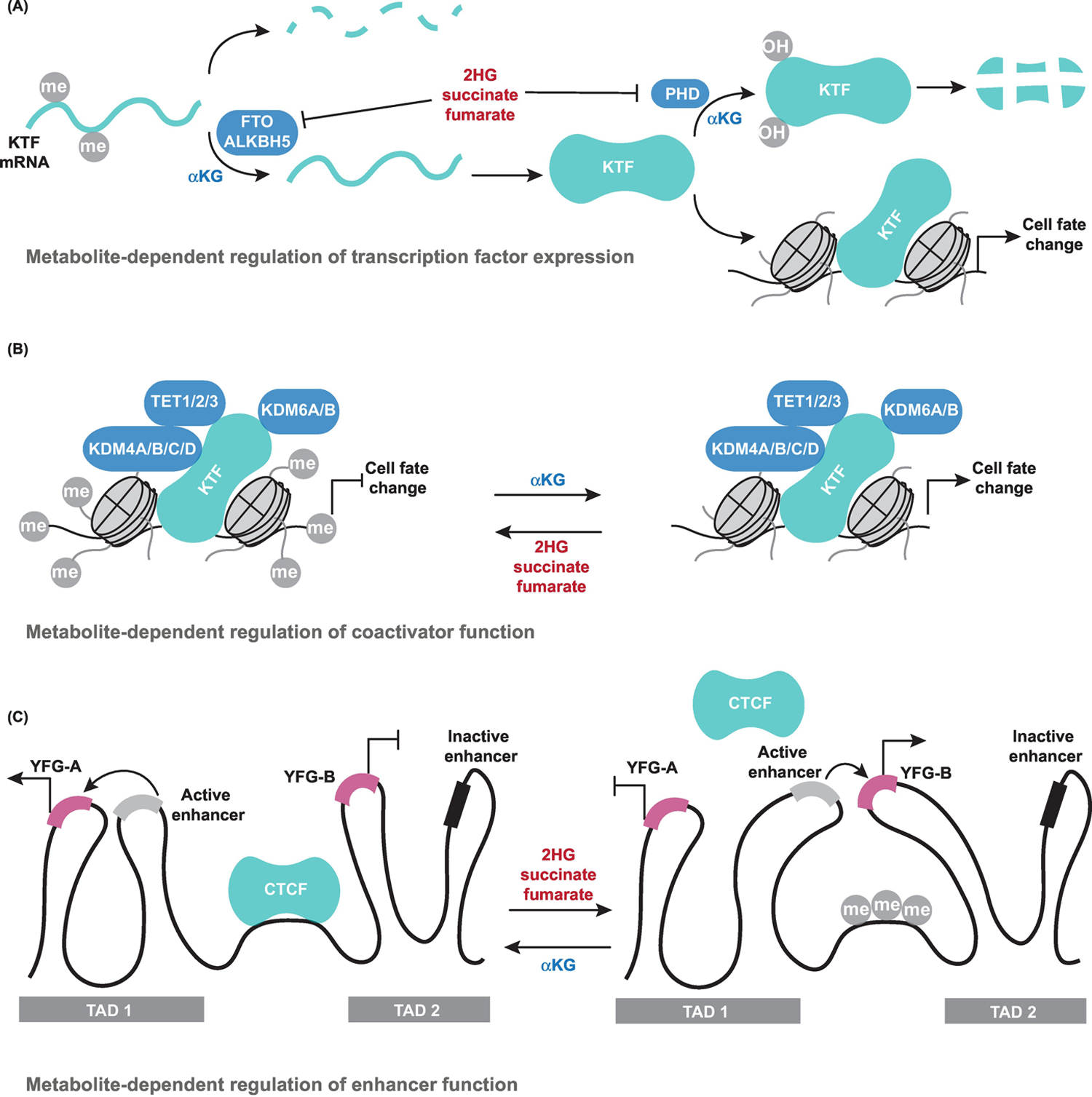
(A) α-Ketoglutarate (αKG)-dependent dioxygenases can directly impact expression of key transcription factors (KTFs) by regulating either mRNA methylation via FTO and ALKBH5 or protein hydroxylation by prolyl hydroxylases (PHDs), each of which trigger target degradation. (B) Metabolites can regulate transcription factor (TF) function by influencing function of transcriptional coactivators, a subset of which are αKG-dependent dioxygenases. Transcription factors recruit coactivators such as ten-eleven translocation (TET) DNA cytosine oxidizing enzymes and Jumonji C-domain lysine demethylases (KDMs) to target loci in order to locally remodel chromatin. (C) Metabolites can affect enhancer function and long-range chromatin interactions by controlling CCCTC-binding factor (CTCF) binding to DNA, which is suppressed by DNA methylation. αKG enforces topologically associated domains (TAD) architecture in cells by facilitating CTCF binding, whereas 2-hydroxyglutarate (2HG), succinate, and fumarate disrupt TAD architecture. An example scenario illustrates how, in the presence of CTCF, a cell type-specific active enhancer drives expression of your favorite gene-A (YFG-A), whereas YFG-B is suppressed. Upon loss of CTCF binding, however, TAD boundaries are disrupted and the active enhancer drives expression of YFG-B.
