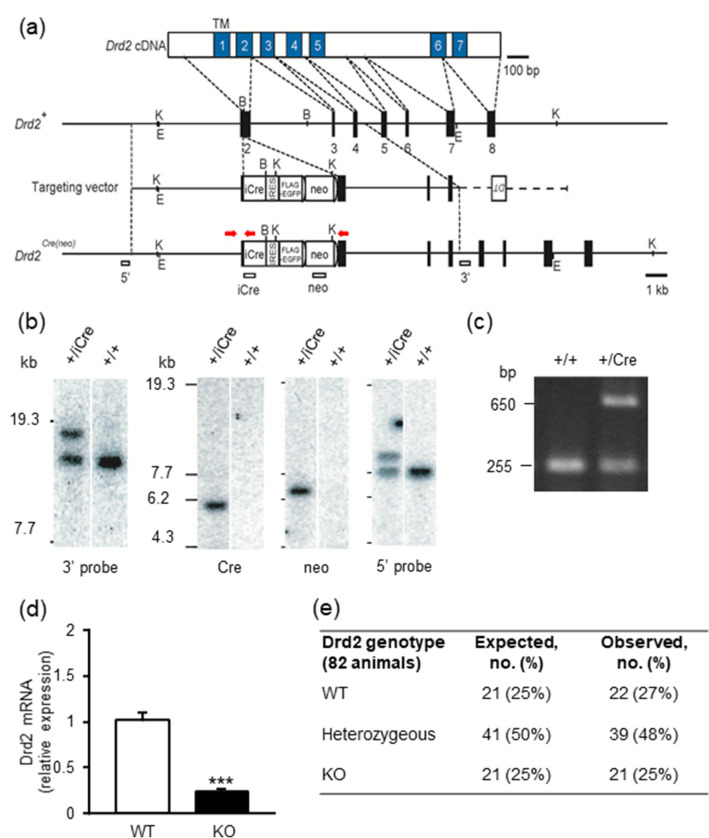
Figure 6.
Generation of the Drd2-Cre mouse line. (a) Schema of Drd2 cDNA, genomic DNA, targeting vector and the targeted genome. Numbered blue boxes in cDNA are transmembrane domains, and black boxes indicate coding exons. The vector was constructed to insert an improved Cre recombinase gene (iCre), followed by an internal ribosome entry site (IRES) and a FLAG-tagged EGFP, into the translational initiation site of the Drd2 gene. Red arrows show primer positions for PCR genotyping. White boxes indicate probes for Southern blot analysis. Two frt sequences (semicircles) are attached to remove the neomycin resistance gene (Neo). B, BamH1; E, EcoT22I; and K, KpnI. (b) Southern blot analysis for genomic DNAs from wild-type (+ / +) and targeted (+ /iCre) ES cells. The first panel from left shows EcoT22I-digested genomic DNA hybridized with 3′ probe, and the other three panels show KpnI-digested DNA hybridized with Cre, BamHI digested DNA, Neo and 5′ probes, respectively. Full-length images are available in Supplementary Fig. 1. (c) Genomic PCR genotyping of Drd2+/iCre using Drd2 forward, reverse and Cre primers. Full-length images are available in Supplementary Fig. 2. (d) Expression of Drd2 mRNA in the hypothalamus of wild-type (WT) mice and Drd2 knockout (KO) mice. Each point represents the mean ± S.E.M. of 6–9 mice. ***P < 0.001 vs. WT group. (e) Observed genotypes of offspring of heterozygous intermatings, in comparison with expected genotypes calculated according to the total number of mice born (82 mice) and the expected Mendelian 1:2:1 ratio.