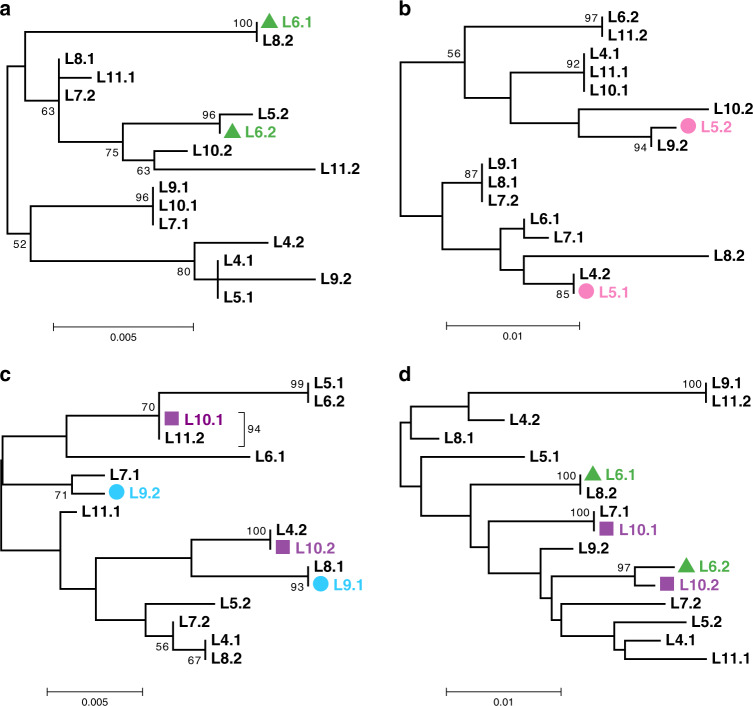
Fig. 5. Phylogenetics of L4-L11 haplotypes indicates interindividual genetic exchange.
Midpoint-rooted phylogenetic trees constructed for four phased segments exhibiting incongruent groupings of individuals with respect to two haplotypes. Overall, in 52 out of 303 tested segments, two haplotypes of at least one individual had their reciprocal closest counterparts in two different individuals (see “Methods” section and Table 1). The displayed phylogenies were constructed for four phased segments located on different contigs in the L1 diploid assembly and spanning 896 (a), 604 (b), 1348 (c), and 1198 (d) base pairs. Trees were built using the maximum likelihood method in PhyML74 under the GTR + G model with 1000 bootstrap replicates. The bootstrap support values ≥50% are shown next to branches. Bootstrap support for the grouping of L10.1 and L11.2 in the panel c is shown next to the bracket. Bootstrap support values are rounded to the nearest integer. Only those SNPs that satisfied all filtering criteria and were simultaneously phased in L4-L11 were used to reconstruct phylogenies (n = 23, 21, 33, and 57 non-singleton SNPs in a–d, respectively); the remaining sites were treated as monomorphic (see “Methods” section). Indices 1 and 2 designate the two haplotypes of a single individual. In these four segments, the two haplotypes of the same individual clustered with haplotypes from two different individuals with high bootstrap support values (≥70%) for L5 (b; pink), L6 (a, d; green), L9 (c; blue), and L10 (c, d; violet). Source data are provided as a Source Data file.