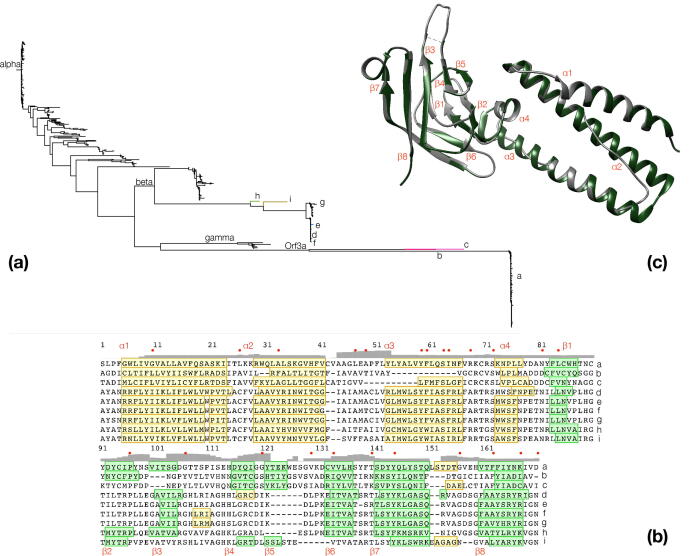
Fig. 2.
Target selection and sequence-structure alignment. (a): A phylogenetic tree depicting relationships of M protein groups and Orf3a within the non-redundant alignment; characters (a-i) signify selected superfamily members (3 from Orf3a, 6 from M protein) for model building by homology, marked by different branch colors. (b): Multiple sequence alignment of targets (‘a-i’, Table S2) as in (a); conserved residues/secondary structure elements are colored and labeled as in Fig. 1; minor alpha-helical segments in some models following α4 are not labeled; grey bar plot signifies RMSD values between models; note that conservation cannot be appreciated in this alignment subset, which is solely used for model building. (c): Orf3a structure alignment between the native structure (green) and model {a} (native structure, excluding the three low-occupancy regions, see also text and Methods); secondary structure elements are labeled as in Fig. 1; this orientation is used elsewhere (Figure S4/S6). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)