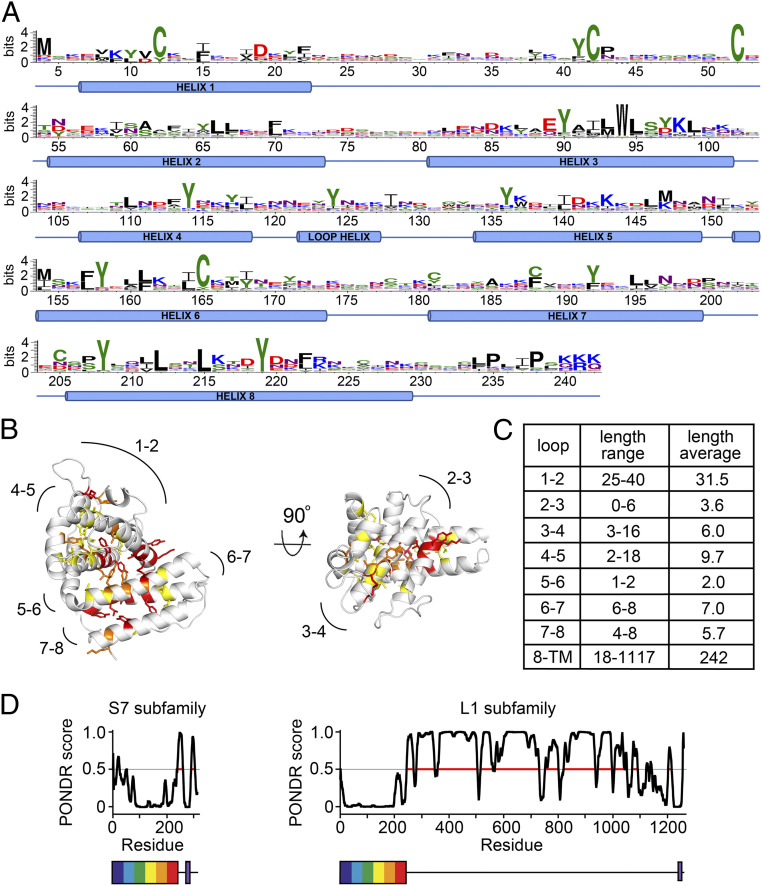
Fig. 2.
Conservation and diversity in the CIR proteins. (A) All CIR sequences from the AS strain of P. chabaudi were aligned against the structure of PCHAS_1200500, and a sequence logo was generated, numbered according to PCHAS_1200500. (B) Residues most chemically conserved across the CIR proteins of the AS strain are shown as sticks. Residues are color-coded by Shannon sequence entropy: 0.75 to 1.0 in yellow, 0.5 to 0.75 in orange, and <0.5 in red. Lower sequence entropy indicates greater conservation of side chain chemical properties. (C) A table of the lengths of the loops and of the linker between the PIR domain fold and the transmembrane helix. (D) Prediction of disorder (PONDR score) for members of the S7 and L1 CIR protein subfamilies, in each case representing the protein closest in sequence to the sequence logo for that protein family, PCHAS_0500200 for the S7 family and PCHAS_0601000 for the L1 family. Prediction of disorder, determined using PONDR, is plotted against residue number. Below the plots are representations of the two proteins, showing the PIR protein domain as a rainbow box and the transmembrane helix in purple.