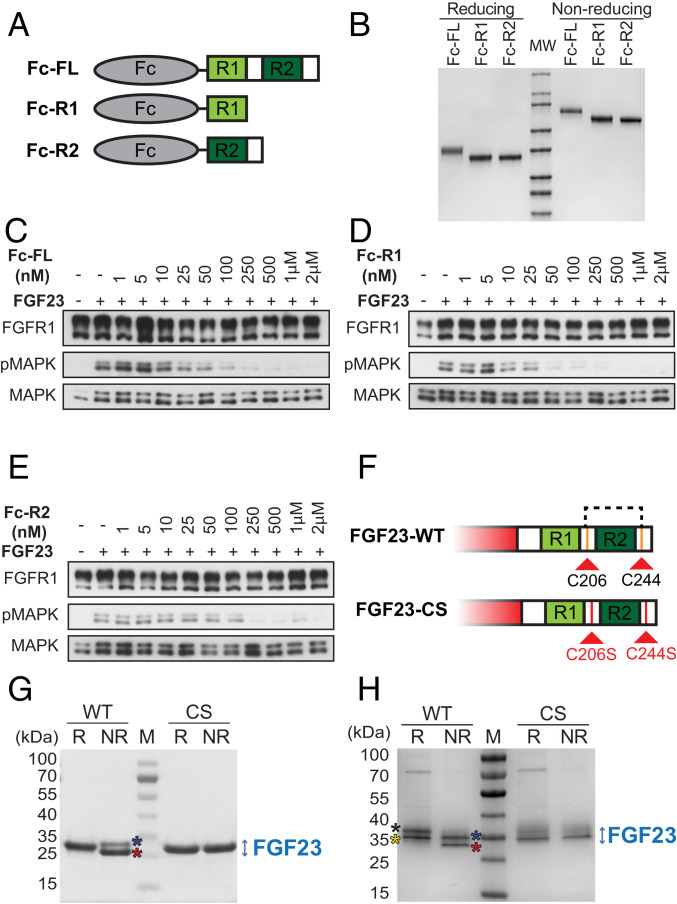
Fig. 3.
Similar inhibition of FGF23-induced stimulation of cells treated with Fc-R2 to cells treated with Fc-FL or Fc-R1 and cysteine residues flanking R2 in FGF23 CT form an intramolecular disulfide bridge. (A and B) Schematic (A) and SDS/PAGE analyzes (B) under reducing (R) or nonreducing (NR) conditions of Fc-FL, Fc-R1, or Fc-R2. The Fc moiety is colored in tan. R1 and R2 are colored in light green and dark green, respectively. (C–E) HEK 293 cells stably expressing FGFR1c and KLA were incubated with increasing concentrations (as indicated) of Fc-FGF23 FL tail (Fc-FL), Fc-R1, or Fc-R2 for 45 min at 37 °C. Cells were then stimulated with FGF23-WT for additional 10 min, and cell lysates were subjected to SDS/PAGE and analyzed for MAPK stimulation by immunoblotting with anti-pMAPK antibodies. Anti-FGFR1 and anti-MAPK antibodies were used as control for protein loading. (F) Schematic of CTs of FGF23-WT and FGF23-CS. R1 and R2 are shown in light green and dark green, respectively. Cysteine residues (C) are highlighted in orange. Serine residues (S) are highlighted in red. (G) SDS/PAGE analyses of FGF23-WT and FGF23-CS mutant expressed in E. coli, under R and NR conditions. FGF23-WT and FGF23-CS were expressed and purified as described in Materials and Methods. While FGF23-CS migrates on SDS/PAGE as a single band under both R and NR conditions, FGF23-WT migrates as two distinct bands under NR conditions. Both proteins were excised from the gel and subjected to mass-spectrometric (MS) analysis. (H) SDS/PAGE analyses of FGF23-WT or FGF23-CS mutant expressed in Expi293F cells under R and NR conditions. Unlike E. coli produced FGF23, mammalian produced FGF23 is O-linked glycosylated. Under R conditions, the upper band (black asterisk) and lower band (yellow asterisk) present an O-linked glycosylated form and a non- or poorly glycosylated form of FGF23, respectively (SI Appendix, Fig. S3B). Under NR conditions, both glycosylated (blue asterisk) and nonglycosylated FGF23 (red asterisk) migrate faster than the reduced proteins on SDS/PAGE due to the formation of intramolecular disulfide bridge. Both FGF23-WT and FGF23-CS (expressed in Expi293F cells) migrate on SDS/PAGE under R and NR conditions as two distinct bands. The upper and lower bands represent O-linked glycosylated and nonglycosylated forms of the ligand, respectively.