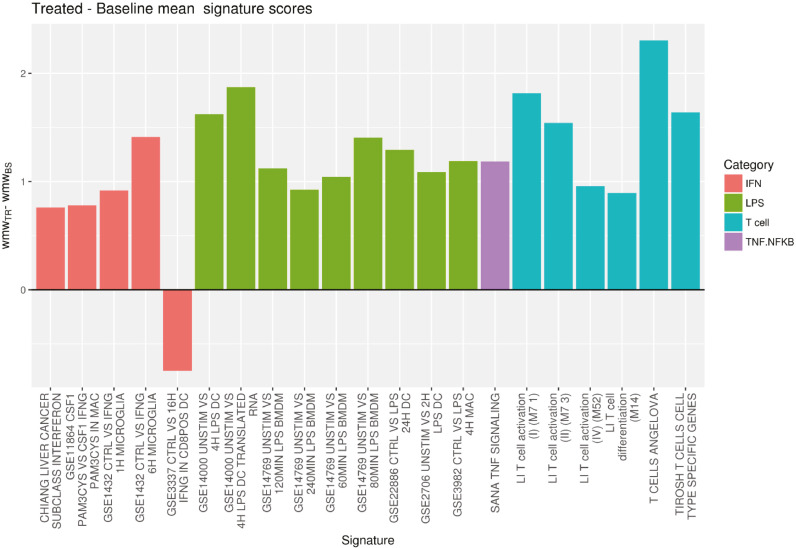
Fig. 6.
Difference between baseline and end-of-treatment signature scores in tumor samples for interferon (IFN), lipopolysaccharide (LPS), T cell and tumor necrosis factor (TNF)/nuclear factor kappa-light-chain-enhancer of activated B (NF-κB) that achieved statistical significance (p < 0.05)a (n = 18). Signatures were divided into several categories that can be an indicator of inflammation (LPS, natural killer [NK], NF-κB and IFN) or of increased expression of genes related to T cells (T-cell signatures from scores from Angelova et al. [28], Li et al. [29] and Tirosh et al. [30]). Individual p-values obtained for the signatures represented above are shown below.
Signature; Category; P value
CHIANG LIVER CANCER SUBCLASS INTERFERON; IFN; 0.028
GSE11864 CSF1 PAM3CYS VS CSF1 IFNG PAM3CYS IN MAC; IFN; 0.034
GSE1432 CTRL VS IFNG 1H MICROGLIA; IFN; 0.039
GSE1432 CTRL VS IFNG 6H MICROGLIA; IFN; 0.046
GSE3337 CTRL VS 16H IFNG IN CD8POS DC; IFN; 0.019
GSE14000 UNSTIM VS 4H LPS DC; LPS; 0.021
GSE14000 UNSTIM VS 4H LPS DC TRANSLATED RNA; LPS; 0.029
GSE14769 UNSTIM VS 120MIN LPS BMDM; LPS; 0.003
GSE14769 UNSTIM VS 240MIN LPS BMDM; LPS; 0.041
GSE14769 UNSTIM VS 60MIN LPS BMDM; LPS; 0.024
GSE14769 UNSTIM VS 80MIN LPS BMDM; LPS; 0.004
GSE22886 CTRL VS LPS 24H DC; LPS; 0.042
GSE2706 UNSTIM VS 2H LPS DC; LPS; 0.013
GSE3982 CTRL VS LPS 4H MAC; LPS; 0.011
SANA TNF SIGNALING; TNF.NFKB; 0.022
LI T cell activation (I) (M7 1); Tcell; 0.031
LI T cell activation (II) (M7 3); Tcell; 0.032
LI T cell activation (IV) (M52); Tcell; 0.046
LI T cell differentiation (M14); Tcell; 0.032
T CELLS ANGELOVA; Tcell; 0.002
TIROSH T CELLS CELL TYPE SPECIFIC GENES; Tcell; 0.044.