FIGURE 3.
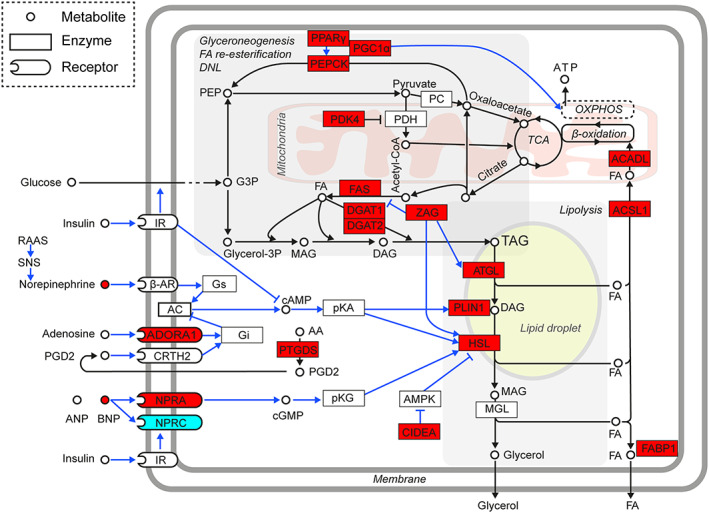
Induction of epicardial adipose tissue (EAT) lipid metabolism linked to cachexia. Gene expression data (Figure 1; and the FC values in Table S2) were collectively analysed to assess activity of the key regulatory and metabolic pathways in EAT adipocytes. Majority of the genes exerted higher mean expression levels (irrespective of the statistical significance of difference) in cachectic as compared with BW‐stable patients; also plasma levels of BNP (Table 2) and adenosine EAT levels (Supporting Information, Dataset S2) were higher in cachectic patients (red colour). Only the NPRC expression in cachectic patients was relatively low (blue colour). Data suggest that stimulation of lipolysis in EAT of HF‐patients by BNP (as well as sympathetic system and RAAS activity; Introduction; not measured here) is further augmented with adipose tissue wasting, in spite of the adaptive antilipolytic response at the PTGDS and ADORA1 gene expression level. Triacylglycerols (TAG) loss due to the increased lipolysis is not fully compensated by lipogenesis, which depends on glyceroneogenesis, de novo fatty acid synthesis (DNL) and fatty acid (FA) re‐esterification. Energy requirements of these anabolic pathways are covered by increased ATP production in mitochondria that combust FA (Figure 7) and other energy fuels (Introduction). Metabolite fluxes and regulatory effects are indicated by black and blue lines, respectively. β‐AR, β‐adrenergic receptor; AA, arachidonic acid; AC, adenylate cyclase; ACADL, long‐chain acyl‐coenzyme A dehydrogenase, also called LCAD; ACSL1, acyl‐CoA synthetase long chain family member 1; ADORA1, adenosine A1 receptor; AMPK, AMP‐activated protein kinase; ANP, A‐type natriuretic peptide; ATGL, adipose triglyceride lipase; BNP, B‐type natriuretic peptide; cAMP, cyclic adenosine monophosphate; cGMP, cyclic guanosine monophosphate; CIDEA, cell death‐inducing DNA fragmentation factor, alpha subunit‐like effector A; CoA, coenzyme A; CRTH2, prostaglandin D2 receptor 2; DAG; diacylglycerol; DGAT1and DAGT2, diacylglycerol O‐acyltransferase 1 and 2, respectively; FABP1, fatty acid binding protein 1; FAS, FA synthase; G3P, glyceraldehyde 3‐phosphate; Gi, G protein subunit alpha i1; Glycerol‐3P, glycerol 3‐phosphate; Gs, guanine nucleotide‐binding protein G(s) subunit alpha; HSL, hormone‐sensitive lipase; IR, insulin receptor; MAG, monoacylglycerol; MGL, monoacylglycerol lipase; NPRA, natriuretic peptide receptor 1; NPRC, natriuretic peptide receptor 3, also called clearance receptor; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase; PDK4, pyruvate dehydrogenase kinase 4; PEP, phosphoenolpyruvate; PEPCK, phosphoenolpyruvate carboxykinase; PGC‐1α, peroxisome proliferative‐activated receptor y coactivator 1α; PGD2, prostaglandin D2; PKA, protein kinase A; PKG, protein kinase G; PLIN1, perilipin 1; PPARγ; peroxisome proliferator activated receptor γ; PTGDS, prostaglandin D2 synthase; RAAS, renin‐angiotensin‐aldosterone system; SNS, sympathetic nervous system; TCA, tricarboxylic acid cycle; ZAG, zinc‐binding alpha‐2‐glycoprotein 1.
