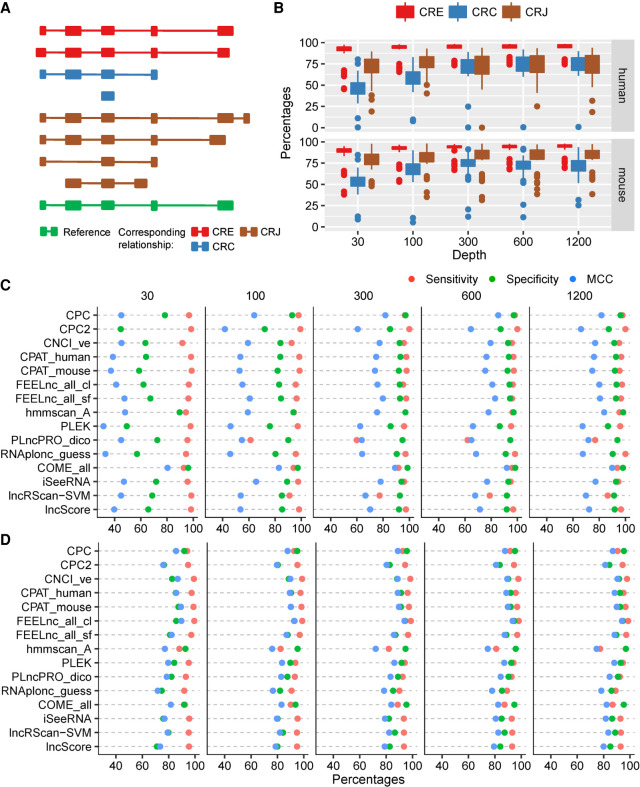
FIGURE 2.
The performance of the representative models on the transcripts assembled from the simulated data. Based on the golden-standard sequence sets (golden_human and golden_mouse), simulated Illumina-sequencing data sets were generated by Polyester with five sequencing depths: 30× (which means that, on average, 30 reads were simulated from each transcript), 100×, 300×, 600×, and 1200×. (A) The relationship between assembled transcripts and its templates. The CRE transcript shares an identical intron chain with its template; the CRC transcript is covered by its template; and the CRJ transcript shares at least one splicing site with its template. (B) Box plots for the MCC of all testing models on the simulated data of different sequencing depths. (C) Performance of representative models on human CRC transcripts. (D) Performance of representative models on human CRJ transcripts.