Figure 1.
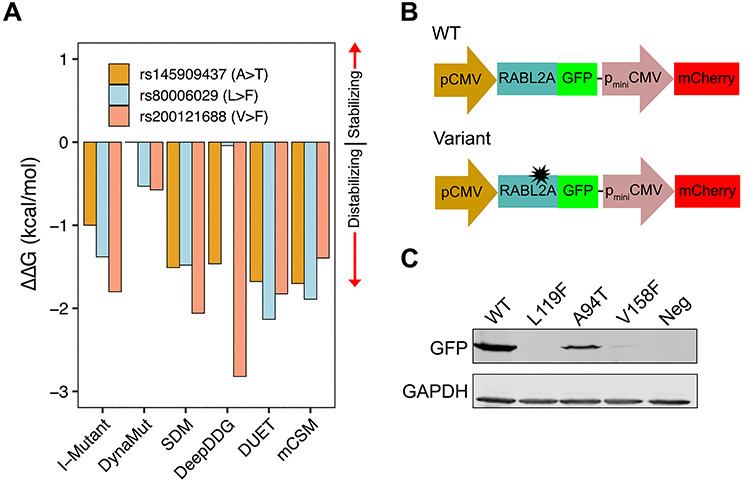
Experimental testing of nsSNPs. (A) Prediction of ΔΔG in p.Ala94Thr, p.Leu119Phe and p.Val158Phe variants by six protein stability prediction algorithms. For I-Mutant, ΔΔG (1 kcal/mol = 4184 J) < −0.5 indicates large decrease of stability, ΔΔG > 0.5 indicates large increase of stability and −0.5 ≤ ΔΔG ≤ 0.5 represents neutral stability. For DynaMut, SDM, DeepDDG, DUET and mCSM, positive ΔΔG indicates stabilization by the specified mutation and negative ΔΔG indicates destabilization. (B) Schematic of the pDEST-DUAL vector with RABL2A WT and variant ORFs used for protein stability assay. (C) Western blots for representative WT and variants detected using α-GFP. α-GAPDH was used as a loading control. Cells transfected with empty plasmid used as negative control.
