Abstract
Ischemic stroke represents one of the leading causes of mortality worldwide and especially in developing countries. It is crucial for finding effective therapeutic targets that protect the brain against ischemic injury. Long noncoding RNAs (lncRNAs) have emerged as major regulators of neurological diseases, and clarifying their roles in cerebral ischemic injury may provide novel targets for the treatment of ischemic stroke. We aimed to investigate the role of lncRNA-XLOC_035088 in middle cerebral artery occlusion (MCAO)-induced rat brain injury and oxygen-glucose deprivation (OGD)-reperfusion treated hippocampal neurons. In our findings, we found that XLOC_035088 expression was significantly upregulated in OGD-reperfusion treated hippocampal neurons and in different brain regions of MCAO-treated rats. XLOC_035088 silencing protected against MCAO-induced ischemic brain injury in vivo and OGD-induced hippocampal neuronal apoptosis in vitro. Intrahippocampal silencing of XLOC_035088 significantly decreased brain XLOC_035088 expression, reduced brain infarct size, and improved neurological function through inhibiting NOTCH1 following derepression of presenilin 2 (PSEN2). Taken together, this study provides evidence that the lncRNA XLOC_035088/PSEN2/Notch1 axis is involved in the pathogenesis of ischemic brain injury, and presents a promising therapeutic route for ischemic stroke.
Keywords: Ischemic stroke, Long noncoding RNA, Middle cerebral artery, Occlusion, XLOC_035088
INTRODUCTION
Ischemic stroke represents one of the leading causes of mortality worldwide and especially in developing countries (1–4), and it is crucial to find effective therapeutic targets that protect the brain against ischemic injury (5, 6). Previous studies have shown that ischemia is followed by many complicated events, such as energy failure, neurotoxicity of excitatory amino acids, inflammatory responses, and depolarization of penumbra and apoptosis, which in turn results in apoptotic and necrotic neuronal death (7–11). Although these mechanisms have been validated in experimental stroke, none of the neuroprotective drugs developed around them have achieved clinical benefit in clinical trials. Further elucidation of the underlying mechanisms of ischemia-induced neuronal death and neurological dysfunction is still required for developing effective therapeutic targets.
Long noncoding RNAs (lncRNAs) are a novel class of mRNA-like transcripts with lengths >200 nt (12). They are involved in epigenetic, transcriptional, and posttranscriptional regulation of diverse biological processes (12–22). Increasing evidence has revealed that mutations and dysregulations of lncRNAs are tightly associated with several human diseases ranging from neurodegenerative diseases to a multitude of cancers (17, 19–25). For example, previous studies have shown that FosDT inhibition significantly improved postischemic motor deficits and reduced infarcted volume (26). Increased expression level of MALAT1 also promotes lung cancer brain metastasis (27). Our preliminary transcriptomic analysis revealed that, compared with the normal brain tissue, there were many differentially expressed lncRNA in the brain tissue of the middle cerebral artery occlusion (MCAO)-induced rat brain injury model. Among them, lncRNA XLOC_035088 was significantly upregulated in the MCAO model group. However, current research on XLOC_035088 has rarely been reported, especially in ischemic stroke.
The presenilin 2 (PSEN2) gene, located on chromosome 1q31-q42, is highly expressed in the cerebellum and hippocampus, and is closely associated with neurodegenerative diseases (26, 27). Previous studies showed that the PSEN2 gene is one of the causative genes of early-onset Alzheimer disease (28). Moreover, PSEN2 is a component of γ-secretase complex, which is an intramembrane-cleaving protease that can cleave the Notch receptors (29). NOTCH1 not only plays an important role in normal cell differentiation, but also regulates the biological processes, such as the occurrence and development of some tumors (29–32). Recent studies have shown that NOTCH1 can be used both as an oncogene and as a tumor suppressor, such as in oral squamous cell carcinoma (33). Clinical studies have shown that NOTCH1 is abnormally activated in colon cancer and participates in the progression of cancer (34). NOTCH1 is also overexpressed in glioma cancer cells, and inhibits proliferation of glioma cells through suppressing the Notch1-Hes-1 signaling pathway (35). Furthermore, NOTCH1 plays an important biological function as a tumor suppressor gene in certain diseases, such as nonsmall cell lung cancer and hepatocellular carcinoma (36, 37). In our preliminary studies, transcriptome analysis indicated that XLOC_035088 and PSEN2 are both dysregulated in MCAO and interacted with each other in the coexpression network, and additional bioinformatics analysis predicted a binding site between XLOC_035088 and PSEN2 3′UTR (Fig. 1A), suggesting that there may be an interaction between XLOC_035088 and PSEN2. However, the role of XLOC_035088/PSEN2/NOTCH1 axis in MCAO-induced rat brain injury in vivo and oxygen-glucose deprivation (OGD)-treated cerebral cortex neuron in vitro is unknown. In-depth study of this potential regulatory pathway could provide important data for the understanding of MCAO.
FIGURE 1.
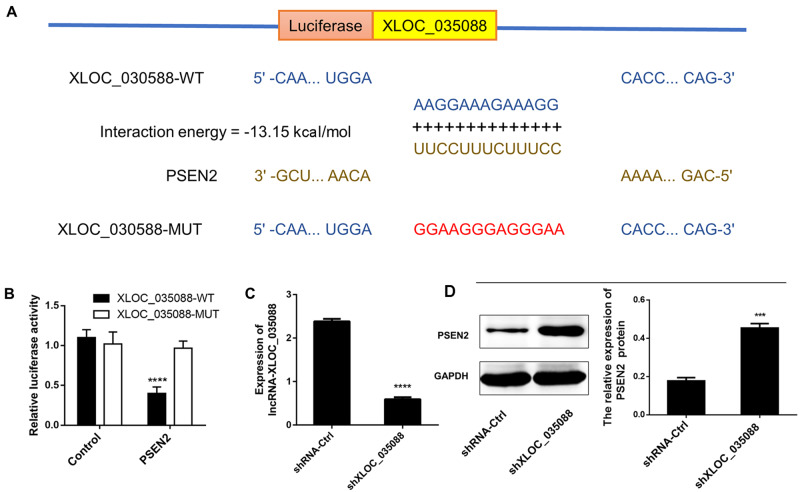
LncRNA-XLOC_035088 is bound to PSEN2. (A) IntaRNA 2.0 predicted 3′-UTR of PSEN2 as a target for XLOC_035088. (B) Wild-type or mutant XLOC_035088 subcloned into luciferase vectors were co-transfected with PSEN2 and the luciferase activities in each group were measured. (C) Detection of mRNA expression levels of XLOC_035088 in normal hippocampus neuron cells transfected with shXLOC_035088 by RT-PCR in vitro study. (D) Detection of protein expression levels of PSEN2 in normal hippocampus neuron cells transfected with shXLOC_035088 by Western blotting in vitro study. ***p < 0.001, ****p < 0.0001 when compared with the control or shRNA ctrl group.
In this study, we investigated the role of XLOC_035088 in MCAO-induced rat brain injury in vivo and OGD-treated cerebral cortex neuron in vitro. XLOC_035088 silencing protects against ischemic brain injury in vivo and neuronal apoptosis in vitro. XLOC_035088 silencing could reduce ischemia-induced brain infarct size and improve neurological function.
MATERIALS AND METHODS
Primary Hippocampus Neuron Cells Culture
All experiments were approved by the Institutional Animal Care Committee and China Council on Animal Care. Sprague-Dawley (SD) rats were purchased from the SLRC Laboratory Animal Company of Shanghai. Primary Hippocampus neuron cultures were established from the cerebral hippocampus of newborn SD rats (<24 hours). Brains from the newborns were cleaned of meninges and blood vessels. The whole cerebral hippocampus was isolated and cells were dissociated in 0.25% trypsin solution for 10 minutes at 37°C. Fetal bovine serum (Gibco, Waltham, MA) was added and the dissociated cells were forced through a mesh. After centrifugation, the hippocampus neuron cells were resuspended and plated in poly-d-lysine (Sigma-Aldrich, St. Louis, MO) coated six-well plates, and grown in Neurobasal medium (Gibco) supplemented with 500 lM glutamine (Gibco) and 2% B27 (Gibco) in a humidified incubator with 5% CO2 at 37°C. To suppress the outgrowth of glial cell populations in primary neuronal cultures, 5 μM cytosine arabinoside ([AraC]; Sigma-Aldrich) was added to inhibit the proliferation of nonneuronal cells at 2 days in vitro and switched to fresh culture medium after 24 hours. Then, the cell culture medium was replaced with fresh culture medium every 4 days. At 14 days in vitro, a near-pure neuron population (>95%) was ready for downstream experiment.
Luciferase Activity Assay
The putative binding sites of lncRNA XLOC_035088 and PSEN2 was predicted by IntaRNA 2.0 (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) (38) with the default parameters (i.e. the number of interactions per RNA pair was 1 and the overlapping interactions in the suboptimal output were allowed overlaps in query only). No lonely base pairs were considered for prediction and no GU base pair was allowed at helix ends. At least 7 base pairs between molecules in the seed area was retained and seeds with GU ends were ignored. The sequences of XLOC_035088 containing PSEN2 binding sites (wild-type [WT]) and mutant not containing PSEN2 binding sites (mutant-type [MUT]) were amplified using PCR, and then subcloned into pGL3 luciferase promoter plasmid (Promega, Madison, WI), respectively, to construct luciferase reporter vectors (XLOC_035088-WT and XLOC_035088-MUT). For luciferase activity assay, the control plasmid and combined plasmid were transfected into hippocampus neuron cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Following the manufacturer's protocols, luciferase assay kit (Promega) was used to measure the luciferase activity after the transfection for 48 hours.
MCAO Animal Model
All animal experiments in this study were carried out in accordance with the NIH Guide for care and use of laboratory animals (NIH Publication No. 80-23). All procedures were approved by the Animal Care and Use Committee of Nanjing Medical University. The rat MCAO model of ischemic stroke was performed. Briefly, 2-month-old male SD rats were anesthetized with an intraperitoneal injection of pentobarbital sodium (60 mg/kg). A blunt dissection was performed under a stereomicroscope to expose the left common carotid artery (CCA), the left external carotid artery (ECA), and the left internal carotid artery (ICA). The CCA was temporarily occluded by a suture. The ECA was permanently sutured as distally as possible. The ICA was sutured distally to the bifurcation. Subsequently, a small incision was performed in the CCA between permanent and temporary sutures. The 5-0 surgical nylon filament was then inserted into the ICA to occlude the origin of the middle cerebral artery (MCA). After 2 hours MCAO, the rats were allowed to recover for 24 hours. In the sham group, all procedures were identical except for inserting an intraluminal filament.
Tissue Preparation
Six, 12, and 24 hours after ischemia-reperfusion, the SD rats (n = 6, each time point) were anesthetized by intraperitoneal injection of 4% chloral hydrate, and the chest was quickly opened to expose the heart. The rats were transcardially perfused with normal saline and after washing away the blood, saline was replaced with 4% paraformaldehyde for perfusion. Subsequently, the brain tissue was taken out, the ipsilateral hippocampus (3–5 mm after the bregma), striatum and cerebral cortex (1–3 mm before the bregma) were isolated according to the stereotaxic map of the rat brain, and placed in 4% paraformaldehyde for 6–8 hours. After transfer to 30% sucrose for dehydration balance and freezing on a freezer with sucrose, and continuously coronal sectioning with a cryostat, each piece had a thickness of ∼10 μm. As a negative control for unilateral brain lesion in the rat model of MCAO, the hippocampus tissue in contralateral hemisphere (nonstroke side of the brain) was also isolated and preserved in the same way.
Neurological Function and Measurement of Infarct Volume
Neurological deficits were scored by a double-blind testing based on a modified Longa EZ test (39), on a scale from 0 to 5 in which 0 represented no deficit, 4 indicated the maximal deficit and 5 indicated mortality at different time points (6, 12, and 24 hours) following MCAO. Results were statistically analyzed using the rank sum test. Brain infarct size was determined at different time points (6, 12, and 24 hours) after the MCAO treatment with 2, 3, 5-triphenyl tetrazolium chloride monohydrate ([TTC]; Sigma-Aldrich) staining. Briefly, the coronal brain slices (2 mm thickness) were cut and immersed into 2% TTC solution for 15 minutes and then fixed in 10% formaldehyde overnight. The caudal and rostral surfaces of each slice were observed. The percentage of infarct area was calculated using Image J software.
OGD-Reperfusion Treatment
Primary Hippocampus neurons were placed in an anaerobic chamber (HERA cell 150, partial oxygen pressure was maintained below 2 mmHg). The medium was replaced with a prewarmed (37°C) glucose-free balanced salt solution. The solution was bubbled with an anaerobic gas mixture (95% N2, 5% CO2) for 30 minutes. Cell cultures subjected to OGD were incubated in the solution at 37°C for different periods to produce oxygen deprivation and then re-oxygenated (returned to the normal aerobic environment). Experimental parameters were assayed at 4 hours after re-oxygenation and repeated 3 times.
Reverse Transcription Quantitative Polymerase Chain Reaction
Total RNAs from the brain tissue (cerebral cortical, striatal and hippocampal regions) and hippocampus neuron cells were isolated using the TRIzol reagent (Invitrogen). They were reversely transcribed by the Superscript III First-Strand Synthesis System (Invitrogen). Reverse transcription quantitative polymerase chain reaction (qRT-PCR) assays were conducted using the Brilliant SYBR Green II qRT-PCR kit (Strategene, La Jolla, CA) according to manufacturer instructions. Each of these reactions involved 10 µL SYBR Green master mix, 2 µL PCR primer mix, and 8-µL diluted cDNA template in a total volume of 20 µL. All PCR assays were conducted in triplicate and threshold cycle numbers were averaged for each sample. The primer sequences used in this experiment are as follows: NOTCH1 forward: 5′-GCCGCCTTTGTGCTTCTGTTC-3′, reverse: 5′-CCGGTGGTCTGTCTGGTCGTC-3′; PSEN2 forward: 5′-GAGGCAGTTTGCCCTGTTTG-3′, reverse: 5′-ATTGTCCCGGCACTCATGTT-3′; lncRNA-XLOC_035088 forward: 5′-CAAAAGCCCAGCAGGACC-3′, reverse: 5′-CTGAAATGATTGCTGGTGGTACTA-3′.
CCK-8 Assay
Cell viability was determined by using the Cell Counting Kit (CCK)-8 assays. In brief, the cerebral cortex neurons were seeded into 96-well plates at a density of 3 × 104 cells/well. After the required treatment, ∼10 µL of CCK-8 solution was added to each well, and then incubated for 2–3 hours. Cell viability was detected using the microplate reader (Spectra-Max M5, Molecular Devices, San Jose, CA).
Apoptosis Assay
The apoptosis of cerebral cortex neurons was detected by FITC Annexin V/PI double staining. The resulting pellets were immediately resuspended in the provided binding buffer and subsequently stained with 5 mL of FITC Annexin V and 5 mL of PI according to the kit's instructions (BD Biosciences, Franklin Lakes, NJ). The mixture was left to incubate at room temperature for 15 minutes, and the cells were analyzed using the FACS system.
Immunofluorescence Staining
The brain slices (10 µm) were fixed in 4% PFA for 15 minutes, blocked in 5% BSA for 0.5 hour, and then incubated with the Psen2 primary antibody (Abcam, Cambridge, UK) overnight at 4°C. The brain slices were washed and incubated with Alexa Fluor 594-conjugated antibody (Abcam) for 3 hours at the room temperature. They were finally mounted and cover slipped with the fluorescent mounting medium. The slices were observed using a fluorescent microscope (Olympus, Tokyo, Japan). The immunofluorescence test was repeated at least 3 independent times.
Construction of Vector and Lentiviral Construct Design
The full-length rat cDNA was cloned and inserted into pcDNA-CMV-MCS-EF1-coGFP vector at EcoRI and XbaI restriction sites. The identity of recombinant plasmid pCDH-PSEN2-GFP was confirmed by PCR and DNA sequencing. Two independent shRNAs specifically targeting XLOC_035088 were designed (GenePharma, Shanghai, China). A scrambled shRNA was used as negative control for lncRNA XLOC_035088-shRNAs. After annealing, XLOC_035088-shRNA fragment was cloned into pLKO.1-TRC vector. The identities of recombinant plasmids pcDNA-PSEN2-GFP and pLKO.1-shRNA-XLOC_035088 were confirmed by PCR and DNA sequencing. Following construction, pcDNA-PSEN2-GFP and pLKO.1-shRNA-XLOC_035088 were transfected into a packaging cell line (293 T). Concentrated viral supernatants were generated by ultracentrifugation and assessed by FACS analysis for GFP on transduced control cells. Hippocampus neuron cells were transfected in Polybrene 6 µg/mL (Sigma-Aldrich) with a high titer of control lentivirus. Other Hippocampus neuron cells were transfected with lentivirus encoding PSEN2 and shRNA-XLOC_035088. For the rat model, a high titer of lentiviral transfection complex (1.38 × 109 pfu/mL) was injected into the lateral ventricle by stereotactic technique at one week before MCAO. Right hippocampus injection coordinates from bregma: 3.5 mm backward, 2.5 mm apart on the right side, 3.0 mm deep.
Cell Transfection
PcDNA3.1 vector (Invitrogen) was used for overexpression. shRNAs for knockdown of specific gene were synthesized by GenePharma. Cells transfection was finished by using the Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s protocol.
Western Blot Analysis
The hippocampus neuron cells or hippocampal tissue were collected and lysed in RIPA lysis buffer (Beyotime, Haimen, China) for 30 minutes in an ice bath. Cells lysates were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA). Every sample was prepared in triplicate and the experiment was repeated at least 3 times. After incubation with blocking buffer (TBS + 0.1% Tween-20 + 5% not fat milk), membranes were probed with primary antibodies (NOTCH1, 1:1000, Abcam; PSEN2, 1:1000, Abcam; GAPDH, 1:1000, Abcam) overnight at 4°C. The membranes were then washed with TBST and incubated with an HRP-conjugated secondary antibody (1:5000, Abcam), and signals were detected with ECL reagent (Millipore).
Statistics Analysis
All data were shown as mean ± SEM. Comparisons between 2 different groups were estimated by Student t test (unpaired, 2-tailed). Comparisons between multiple groups were estimated by ANOVA or repeated-measures ANOVA analyses; p < 0.05 was considered statistically significant.
RESULTS
XLOC_035088 Is a Negative Regulator of PSEN2
To verify the interaction between XLOC_035088 and PSEN2 detected in the preliminary studies, IntaRNA 2.0 was used for predicting the potential binding site. A unique binding site with 13 base pairs between 3′-UTR of PSEN2 and XLOC_035088 was predicted, and the energy score of interaction was –13.15 kcal/mol (Fig. 1A). Subsequently, their interaction was proved by luciferase activity assay in hippocampus neuron cells in vitro. Results showed that the expression level of PSEN2 was negatively correlated with luciferase activity when transfected with XLOC_035088-WT (p < 0.01). However, there was no significant difference in response to XLOC_035088-MUT group (Fig. 1B). Moreover, the regulatory relationship between XLOC_035088 and PSEN2 was further verified by silencing XLOC_035088. We first verified that shXLOC_035088 transfection significantly reduced the expression level of XLOC_035088 in normal hippocampus neuron cells by RT-PCR (Fig. 1C), and the effect of XLOC_035088 on PSEN2 expression was measured in normal hippocampus neuron cells (no ischemic stroke) using Western blot after transfection for 72 hours. It was found that silencing of XLOC_035088 significantly upregulated the expression of PSEN2, indicating XLOC_035088 is a negative regulator of PSEN2 (Fig. 1D).
Increased Expression of XLOC_035088 and NOTCH1 and Downregulation of PSEN2 Were Recorded in MCAO Rat Model and OGD-Treated Hippocampus Cells
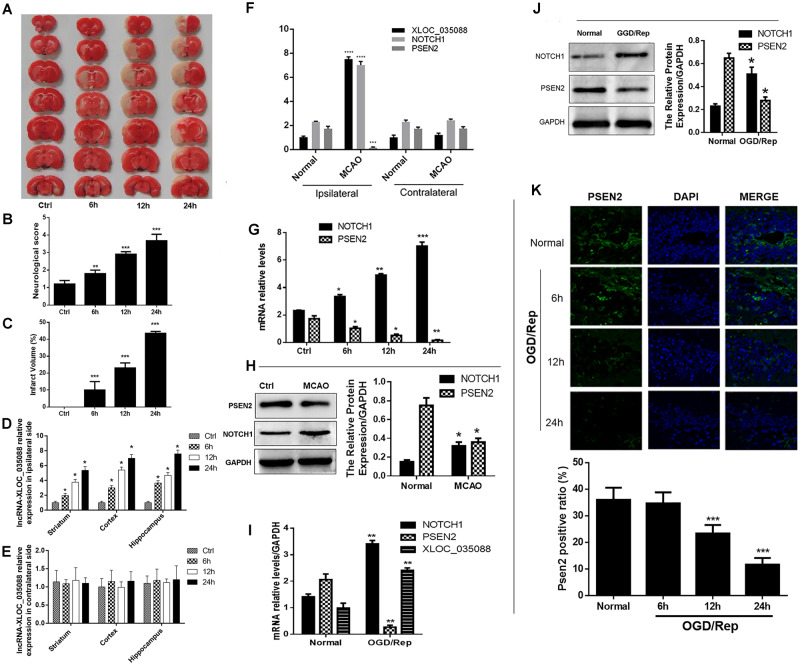
In order to explore the effects of lncRNA-XLOC_035088 on brain injury, we first established a rat MCAO model. The TTC staining and Neurological Severity Scale indicated that the score of neurological deficits and brain infarct volume were markedly increased following MCAO/Rep after 6, 12, and 24 hours compared with the control group (Fig. 2A–C). We next detected the expression of XLOC_035088 in the ipsilateral (injured hemisphere) and contralateral sides of brain tissue at 6, 12, and 24 hours after MCAO/Rep and found that XLOC_035088 was significantly upregulated in the cerebral cortical, striatal and hippocampal region from ipsilateral side compared with that in control group in a time-dependent manner (Fig. 2D). However, no significant difference was found at 3 different time points in the cerebral cortical, striatal and hippocampal region from contralateral side when compared with that in control group (Fig. 2E). Since the expression level of XLOC_035088 was higher in the hippocampus compared with the other 2 regions, hippocampus was used as the sampling region for measuring brain injury indicators in downstream experiments. Compared with the negative control (i.e. hippocampus from the nonstroke side of the brain), there was no significant difference in the expression levels of lncRNA XLOC_035088, NOTCH1 and PSEN2 in the contralateral hemisphere between MCAO and normal groups 24 h after MCAO treatment (Fig. 2F). These results further confirmed that changes in the ipsilateral hemisphere were specific responses to brain injury (Fig. 2D–F). Subsequently, since we aimed to investigate the possible functional relationship between XLOC_035088, PSEN2 and Notch1 in brain injury, we equally determined the expression levels of PSEN2 and Notch1 in the hippocampus of MCAO rats. The qRT-PCR and Western blotting analyses showed that, similarly to XLOC_035088, the expression of NOTCH1 was significantly increased in the rat hippocampus (Fig. 2G, H) while PSEN2 (Fig. 2G, H) was downregulated following MCAO. Similarly, in vitro studies indicated that OGD/Rep treatment led to increased expressions of XLOC_035088 and NOTCH1 but decreased the expression of PSEN2 as evaluated by qRT-PCR (Fig. 2I). Western blotting analysis showed that the expression of PSEN2 was decreased while Notch1 was upregulated by OGD/Rep treatment (Fig. 2J). The decreased expression of PSEN2 was confirmed by Immunofluorescence results (Fig. 2K). These results indicated that the upregulation of XLOC_035088 and NOTCH1 and the downregulation of PSEN2 are probably involved in brain injury induced by MCAO/Rep.
FIGURE 2.
Ischemic injury increased the expressions of XLOC_035088 and NOTCH1 and downregulated PSEN2 in vivo and in vitro. (A) TTC staining of brain sections obtained before and after MCAO-induction. (B) Evaluation of neurological score with and without MCAO (n = 8, each group). (C) Assessment of infarct volume with and without MCAO induction (n = 8, each group). (D) Expression of lncRNA-XLOC_035088 in different brain regions from ipsilateral side at different time points (6, 12, and 24 hours) after MCAO (n = 3 for each time point). (E) Expression of lncRNA-XLOC_035088 in different brain regions from contralateral side at different time points (6, 12, and 24 hours) after MCAO (n = 3 for each time point). (F) Detection of expression levels of lncRNA XLOC_035088, NOTCH1 and PSEN2 in the ipsilateral or contralateral hemisphere in normal and MCAO groups 24 hours after MCAO treatment by RT-PCR. (G) Detection of mRNA expression levels of NOTCH1 and PSEN2 in the hippocampus at different time points (6, 12, and 24 hours) after MCAO by RT-PCR (n = 3 for each time point). (H) Detection of protein expression levels of NOTCH1 and PSEN2 in the hippocampus at different time points (6, 12, and 24 hours) after MCAO by Western blotting (n = 3 for each time point). (I) Detection of mRNA expression levels of NOTCH1, PSEN2, and lncRNA-XLOC_035088 in the hippocampal neurons at 24 hours after OGD/reperfusion by RT-PCR. (J) Detection of protein expression levels of NOTCH1 and PSEN2 in the hippocampal neurons at 24 hours after OGD/reperfusion by Western blotting. (K) Detection of PSEN2 expression in the hippocampal neurons at different time points (6, 12, and 24 hours) after OGD/reperfusion by immunofluorescence. *p < 0.05, **p < 0.01, ***p < 0.001 when compared with ctrl group for the in vivo study or normal group for the in vitro study.
Silencing of XLOC_035088 Suppressed Ischemic Injury and Apoptosis In Vivo and In Vitro
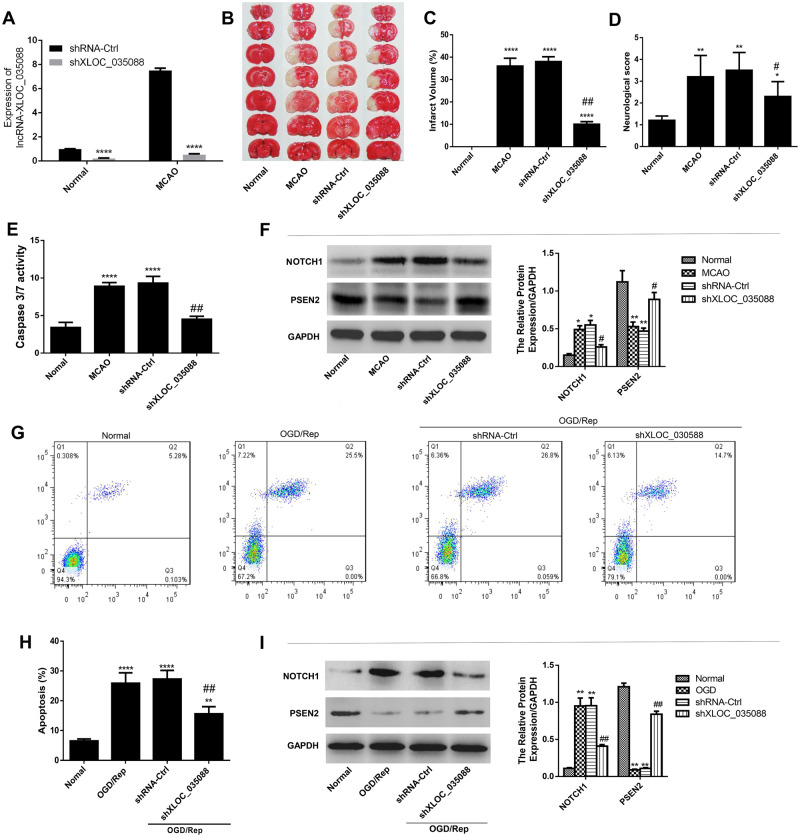
To explore the biological roles of XLOC_035088 in ischemic stroke, we first verified that shXLOC_035088 transfection significantly reduced the expression level of XLOC_035088 both in normal and MCAO model groups by RT-PCR (Fig. 3A). The TTC staining results exhibited that MCAO produced an infarct compared with the control group while XLOC_035088 silencing led to a significant decrease in infarct size compared with the MCAO and sh-control groups (n = 8 per group; Fig. 3B, C). Neurological outcomes data showed that silencing of XLOC_035088 significantly improved long-term neurological outcomes after MCAO compared with sh-control group (Fig. 3D). In addition, caspase-3/7 activity assay indicated that MCAO treatment markedly induced the caspase-3/7 activity in the hippocampus (Fig. 3E) while XLOC_035088 silencing noticeably repressed the increase of caspase-3/7 activity induced by MCAO, suggesting that XLOC_035088 silencing suppressed apoptosis induced by MCAO in vivo. Moreover, silencing of XLOC_035088 significantly downregulated the expression of NOTCH1 and upregulated the expression of PSEN2 compared with MCAO and shRNA-ctrl groups (Fig. 3F).
FIGURE 3.
Silencing of XLOC_035088 suppressed ischemic injury and apoptosis in vivo and in vitro. (A) Detection of expression level of XLOC_035088 after shXLOC_035088 transfection both in normal and MCAO model groups by RT-PCR. (B) TTC staining of brain sections obtained with and without MCAO-induction in rats transfected with shXLOC_035088. (C) Evaluation of neurological score with and without MCAO (n = 8, each group). (D) Assessment of infarct volume with and without MCAO induction (n = 8, each group). (E) Effect of XLOC_035088 silencing on caspase 3/7 activity in rat hippocampus treated with or without MCAO. (F) Detection of protein expression levels of NOTCH1 and PSEN2 in the hippocampus with and without MCAO by Western blotting in rats transfected with shXLOC_035088. (G, H) Detection of cell apoptosis by flow cytometry. (I) Detection of protein expression levels of NOTCH1 and PSEN2 in the hippocampal neurons at 24 hours after OGD/reperfusion by Western blotting. *p < 0.05, **p < 0.01, ****p < 0.0001 when compared with normal group. #p < 0.05, ##p < 0.01 when compared with shRNA ctrl group.
The effect of XLOC_035088 silencing on ischemic injury was also evaluated in vitro. Hippocampal cells were transfected with sh-XLOC_035088 or sh-control prior to OGD/Rep. Flow cytometry analysis indicated that treatment with OGD/Rep significantly induced the apoptosis of hippocampal cells and silencing of XLOC_035088 reversed this effect (Fig. 3G, H). Additionally, we found that silencing of XLOC_035088 repressed OGD/Rep-induced expression of NOTCH1 but reverted the inhibitory effect of OGD/Rep on PSEN2 expression (Fig. 3I). Taken together, silencing of XLOC_035088 suppresses ischemic injury and apoptosis in vivo and in vitro probably via regulating the NOTCH1/PSEN2 axis.
PSEN2 Overexpression Mimicked the Inhibitory Effect of XLOC_035088 Silencing on Ischemic Injury and Apoptosis In Vivo and In Vitro
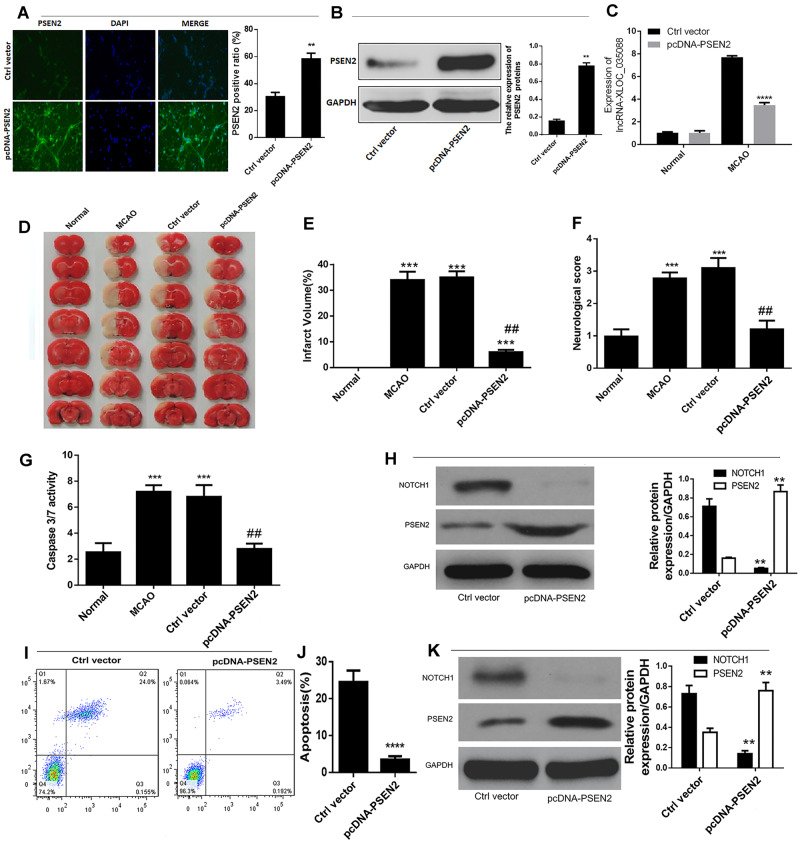
To further understand the interaction between XLOC_035088 and PSEN2 and confirm the impact of PSEN2 on Notch signaling pathway, we first detected the transfection efficiency of pcDNA-PSEN2 by immunofluorescence and Western blotting (Fig. 4A, B) and found that MCAO rats treated with pcDNA-PSEN2 significantly downregulated the expression level of XLOC_035088 (Fig. 4C). Analogously to the inhibitory effect of XLOC_035088 silencing, pcDNA-PSEN2 vector or control vector were injected into the lateral ventricle of rats prior to MCAO (n = 8 per group). Hippocampal neurons were transfected with PSEN2 overexpression vector or empty vector prior to OGD/Rep. As shown in Figure 4D and E, PSEN2 overexpression led to a dramatic reduction of MCAO-induced infarct volume of ischemic brain. Furthermore, PSEN2 overexpression mitigated MCAO-induced neurological deficit of rats (Fig. 4F). Additionally, PSEN2 overexpression significantly reduced the expression of NOTCH1 and caspase 3/7 activity in hippocampal of MCAO-induced rat (Fig. 4G, H). As demonstrated by flow cytometry analysis, PSEN2 overexpression significantly repressed apoptosis of hippocampal neuron cells after OGD/Rep in comparison with control vector-transfected cells (Fig. 4I, J). Additionally, the expression level of NOTCH1 was remarkably decreased in PSEN2-overexpressing hippocampal neurons after OGD/Rep (Fig. 4K). These effects were similar to the effect of XLOC_035088 silencing. Taken together, these data suggested that XLOC_035088 silencing protects against ischemic stroke by promoting the expression of PSEN2 through inhibiting the NOTCH1 signaling.
FIGURE 4.
Overexpression of PSEN2 suppressed ischemic injury and apoptosis in vivo and in vitro. (A, B) Detection of transfection efficiency of pcDNA-PSEN2 by immunofluorescence and Western blotting. (C) Detection of expression level of XLOC_035088 after pcDNA-PSEN2 transfection both in normal and MCAO model groups by RT-PCR. (D) TTC staining of brain sections obtained with and without MCAO-induction in rats transfected with PSEN2 overexpression vector. (E) Assessment of infarct volume with and without MCAO induction (n = 8, each group). (F) Evaluation of neurological score with and without MCAO (n = 8, each group). (G) Detection of protein expression levels of NOTCH1 and PSEN2 in the hippocampus after MCAO by Western blotting in rats transfected with shXLOC_035088. (H) Determination of capase3/7 activity in the hippocampus following MCAO. (I, J) Detection of cell apoptosis by flow cytometry 24 hours following OGD/Repd. (K) Detection of protein expression levels of NOTCH1 and PSEN2 in the hippocampal neurons 24 hours after OGD/reperfusion by Western blotting. **p < 0.01, ****p < 0.0001 when compared with normal group or ctrl vector. #p < 0.05, ##p < 0.01 when compared with ctrl vector or MCAO group.
DISCUSSION
Dysregulation of lncRNAs have been found in a broad range of human diseases, including cancers, diabetes, and heart diseases (40, 41). In this study, we first demonstrated that lncRNA XLOC_035088 functioned as a negative regulator for PSEN2 to altered levels of NOTCH1in the Notch signaling pathway, contributing to ischemic stroke injury in MCAO-induced rat brain in vivo and OGD/Rep-induced hippocampal neuron in vitro.
LncRNAs are important regulators of significant biological processes and have been incriminated in the development of human brain diseases. This study demonstrates for the first time that the expression of XLOC_035088 is increased in MCAO-treated rat brain and OGD-reperfusion treated hippocampal cells. It is also the first to show that XLOC_035088 silencing has a potential to protect hippocampal neurons from ischemia-induced brain injury. Previous studies showed that nerve cells are most sensitive to ischemia (42). Even in neural cells, different types of nerve cells have different sensitivities to ischemia and are also related to the location of nerve cells. The sensitivity of nerve cells in different parts to ischemia from largest to the smallest were hippocampus, striatum and cortex, and ischemia had the greatest damage to the hippocampus (43). Thus, neuronal cell injury is the key outcome of ischemia-induced stroke (44–46). Previous studies have indicated that lncRNAs play fundamental functions in the regulation of neuronal cell function. For instance, previous studies have indicated that lncRNA FosDT is acutely upregulated and induces ischemic brain injury by regulating the REST-associated chromatin-modifying proteins (40).
During ischemic brain injury, neuronal cells undergo various pathophysiological changes, such as apoptosis (47–49). In conformity with previous works (47–49), we found increased apoptosis of hippocampal neurons during ischemic injury in both in vivo and in vitro. Moreover, our present work indicated that XLOC_035088 silencing prevented cell apoptosis. These data are the first to describe a functional role of XLOC_035088 in the regulation of neuronal apoptosis, thus XLOC_035088 will be a potent therapeutic target for ischemic stroke.
The Notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms. Altered levels of NOTCH1 have been detected in diverse brain pathological events and upregulation of NOTCH1 is known to promote neuronal apoptosis (50–54). Ischemic conditions lead to a marked increase in NOTCH1, which may affect cell viability, proliferation and apoptosis and promote ischemic stroke. In the present study we found that XLOC_035088 and NOTCH1 expression were both upregulated by ischemic injury and that silencing of XLOC_035088 was followed by decreased expression of NOTCH1. This suggested that the mechanism underlying the effect of XLOC_035088 on the ischemic injury may be exerted trough NOTCH1 signaling.
Presenilins (PSENs) are known as regulators of amyloid precursor protein (APP) processing by modulating the activity of the APP-cleaving enzyme γ-secretase, and mutations of these proteins are responsible for Alzheimer disease (55–58), but their role in ischemic injury has not been well elucidated. Additionally, previous studies have indicated that the PSENs are implicated in the cleaving protease required in the Notch signaling, especially NOTCH1 (59). In this study, we demonstrated that overexpression of PSEN2 significantly reduced the effect of ischemic injury, which similar to the data obtained with XLOC_035088 silencing. In addition, since XLOC_035088 silencing was followed by increased expression of PSEN2, we conclude that XLOC_035088 promotes ischemic injury by inhibiting PSEN2 expression. In addition, we also found that PSEN2 overexpression was followed by decreased expression of NOTCH1, which was positively regulated by XLOC_035088. Thus, we infer that XLOC_035088/PSEN2/Notch1 axis is a regulatory pathway in ischemic injury.
Our study presents some limitations. Indeed, the interpretations of both cell culture and animal model paradigm results need to be mindful that Notch1 and PSENs are expressed in neuronal cells, glia, and invading immune/inflammatory cells in the MCAO model with infarct. Thus, for both in vivo and in vitro models of ischemic stroke, we need to exclude interference from nonneuronal cells, such as glia, immune/inflammatory cell lines and other blood-derived cells to verify the effects on neuronal cells more accurately. In addition, our work was done out of the context of PSEN2/PSEN1 function as catalytic subunits of γ-secretase and the data on Notch1 were not related to its cleavage by γ-secretase or any of its other 70-some substrates, including APP. For this reason, we plan to further verify the interaction between lncRNA XLOC_035088 and PSEN1, and to clarify the role of XLOC_035088 in the network composed of γ-secretase and its protein substrates, such as Notch receptors and APP in our future experiments. Moreover, we found that shRNA causes the reduction of XLOC_035088 in normal tissue as well, so we need to further explore the effect of XLOC_035088 downregulation on the function of nonstroke tissues in rats in our future studies.
In conclusion, this study provides evidence that XLOC_035088 is upregulated and involved in ischemic brain injury by promoting the expression of PSEN2 through inhibiting the NOTCH1 signaling. Thus, this study provides novel insights that XLOC_035088/PSEN2/Notch1 axis could be a therapeutic target for ischemic stroke.
This study was funded by “Key disciplines of medicine in Yangpu District Year 2020-2022 (Class A)” (Award No. YP19ZA08 to H.W.) and Science Research Project “The regulatory role of lncRNA-miRNA-mRNA network in the prevention and treatment of cognitive impairment after stroke by hyperbaric oxygen therapy” supported by Shanghai Health Committee (Award No. 201940155 to H.W.).
The authors have no duality or conflicts of interest to declare.
REFERENCES
- 1. Behrouz R. The risk of ischemic stroke in major rheumatic disorders. J Neuroimmunol 2014;277:1–5 [DOI] [PubMed] [Google Scholar]
- 2. Hao L, Zou Z, Tian H, et al. Stem cell-based therapies for ischemic stroke. Biomed Res Int 2014;2014:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nave AH, Lange KS, Leonards CO, et al. Lipoprotein (a) as a risk factor for ischemic stroke: A meta-analysis. Atherosclerosis 2015;242:496–503 [DOI] [PubMed] [Google Scholar]
- 4. Ni X, Zhang J.. Association between 9p21 genomic markers and ischemic stroke risk: Evidence based on 21 studies. PLoS One 2014;9:e90255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Regenhardt RW, Das AS, Stapleton CJ, et al. Blood pressure and penumbral sustenance in stroke from large vessel occlusion. Front Neurol 2017;8:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel RAG, Mcmullen PW.. Neuroprotection in the treatment of acute ischemic stroke. Prog Cardiovasc Dis 2017;59:542–8 [DOI] [PubMed] [Google Scholar]
- 7. Ahmad M, Dar NJ, Bhat ZS, et al. Inflammation in ischemic stroke: Mechanisms, consequences and possible drug targets. CNS Neurol Disord Drug Targets 2014;13:1378–96 [DOI] [PubMed] [Google Scholar]
- 8. Gonzalez-Moreno EI, Camara-Lemarroy CR, Gonzalez-Gonzalez JG, et al. Glycemic variability and acute ischemic stroke: The missing link? Transl Stroke Res 2014;5:638–46. [DOI] [PubMed] [Google Scholar]
- 9. Jin R, Liu L, Zhang S, et al. Role of inflammation and its mediators in acute ischemic stroke. J Cardiovasc Trans Res 2013;6:834–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siniscalchi A, Gallelli L, Malferrari G, et al. Cerebral stroke injury: The role of cytokines and brain inflammation. J Basic Clin Physiol Pharmacol 2014;25:131–7 [DOI] [PubMed] [Google Scholar]
- 11. Turtzo LC, Li J, Persky R, et al. Deletion of macrophage migration inhibitory factor worsens stroke outcome in female mice. Neurobiol Dis 2013;54:421–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu Z, Liu X, Liu L, et al. Regulation of lncRNA expression. Cell Mol Biol Lett 2014;19:561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carlson HL, Quinn JJ, Yang YW, et al. LncRNA-HIT functions as an epigenetic regulator of chondrogenesis through its recruitment of p100/CBP complexes. PLoS Genet 2015;11:e1005680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res 2015;35:1385–8 [PubMed] [Google Scholar]
- 15. Lukas J, Altmeyer M.. A lncRNA to repair DNA. EMBO Rep 2015;16:1413–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang K, Liu CY, Zhou LY, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun 2015;6:6779. [DOI] [PubMed] [Google Scholar]
- 17. Yan B, Yao J, Liu JY, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 2015;116:1143–56 [DOI] [PubMed] [Google Scholar]
- 18. Yin DD, Zhang EB, You LH, et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic beta cells. Cell Physiol Biochem 2015;35:1892–904 [DOI] [PubMed] [Google Scholar]
- 19. Riva P, Ratti A, Venturin M.. The long non-coding RNAs in neurodegenerative diseases: Novel mechanisms of pathogenesis. Curr Alzheimer Res 2016;13:1219–31 [DOI] [PubMed] [Google Scholar]
- 20. Wu P, Zuo X, Deng H, et al. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull 2013;97:69–80 [DOI] [PubMed] [Google Scholar]
- 21. Martens-Uzunova ES, Bottcher R, Croce CM, et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 2014;65:1140–51 [DOI] [PubMed] [Google Scholar]
- 22. Zhang A, Xu M, Mo YY.. Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol 2014;6:181–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takahashi K, Yan I, Haga H, et al. Long noncoding RNA in liver diseases. Hepatology 2014;60:744–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uchida S, Dimmeler S.. Long noncoding RNAs in cardiovascular diseases. Circ Res 2015;116:737–50 [DOI] [PubMed] [Google Scholar]
- 25. Yang X, Gao L, Guo X, et al. A network based method for analysis of lncRNA-disease associations and prediction of lncRNAs implicated in diseases. PLoS One 2014;9:e87797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature 1995;376:775–8 [DOI] [PubMed] [Google Scholar]
- 27. Levy-Lahad E, Wasco W, Poorkaj P, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science (New York, NY) 1995;269:973–7 [DOI] [PubMed] [Google Scholar]
- 28. Rogaeva E. The solved and unsolved mysteries of the genetics of early-onset Alzheimer’s disease. Neuromol Med 2002;2:1–10. [DOI] [PubMed] [Google Scholar]
- 29. Lino MM, Merlo A, Boulay JL.. Notch signaling in glioblastoma: A developmental drug target? BMC Med 2010;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miele L. Notch signaling. Clin Cancer Res 2006;12:1074–9 [DOI] [PubMed] [Google Scholar]
- 31. Yoshida R, Ito T, Hassan WA, et al. Notch1 in oral squamous cell carcinoma. Histol Histopathol 2016;32:11821. [DOI] [PubMed] [Google Scholar]
- 32. Roy M, Pear WS, Aster JC.. The multifaceted role of Notch in cancer. Curr Opin Genet Dev 2007;17:52–9 [DOI] [PubMed] [Google Scholar]
- 33. Yoshida R, Ito T, Hassan WA.. Notch1 in oral squamous cell carcinoma. Histol Histopath 2016;32:11821. [DOI] [PubMed] [Google Scholar]
- 34. Meng RD, Shelton CC, Li Y-M, et al. gamma-Secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res 2009;69:573–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yao J, Zheng K, Li C, et al. Interference of Notch1 inhibits the growth of glioma cancer cells by inducing cell autophagy and down-regulation of Notch1-Hes-1 signaling pathway. Med Oncol 2015;32:174. [DOI] [PubMed] [Google Scholar]
- 36. Yang YL, Jablons D, You L.. An alternative way to initiate Notch1 signaling in non-small cell lung cancer. Transl Lung Cancer Res 2014;3:238–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hany H, Shalaby A, Al Kashef W, et al. Evaluation of the role of Notch1 expression in hepatic carcinogenesis with clinico-pathological correlation. Pathology 2018;50:730–6 [DOI] [PubMed] [Google Scholar]
- 38. Mann M, Wright PR, Backofen R.. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res 2017;45:W435–w9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu G, Wang T, Wang T, et al. Effects of apoptosis-related proteins caspase-3, Bax and Bcl-2 on cerebral ischemia rats. Biomed Rep 2013;1:861–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mehta SL, Kim T, Vemuganti R.. Long noncoding RNA FosDT promotes ischemic brain injury by interacting with REST-associated chromatin-modifying proteins. J Neurosci 2015;35:16443–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen L, Chen L, Wang Y, et al. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol 2015;121:101–8 [DOI] [PubMed] [Google Scholar]
- 42. Butterfield JD Jr, Mcgraw CP.. Free radical pathology. Stroke 1978;9:443–5 [DOI] [PubMed] [Google Scholar]
- 43. Zheng YQ, Liu JX, Li XZ, et al. RNA interference-mediated downregulation of Beclin1 attenuates cerebral ischemic injury in rats. Acta Pharmacol Sin 2009;30:919–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benjelloun N, Joly LM, Palmier B, et al. Apoptotic mitochondrial pathway in neurones and astrocytes after neonatal hypoxia-ischaemia in the rat brain. Neuropathol Appl Neurobiol 2003;29:350–60 [DOI] [PubMed] [Google Scholar]
- 45. Patel AR, Ritzel R, McCullough LD, et al. Microglia and ischemic stroke: A double-edged sword. Int J Physiol Pathophysiol Pharmacol 2013;5:73–90 [PMC free article] [PubMed] [Google Scholar]
- 46. Wang P, Xu TY, Wei K, et al. ARRB1/beta-arrestin-1 mediates neuroprotection through coordination of BECN1-dependent autophagy in cerebral ischemia. Autophagy 2014;10:1535–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lv H, Wang L, Shen J, et al. Salvianolic acid B attenuates apoptosis and inflammation via SIRT1 activation in experimental stroke rats. Brain Res Bull 2015;115:30–6 [DOI] [PubMed] [Google Scholar]
- 48. Sun Y, Gui H, Li Q, et al. MicroRNA-124 protects neurons against apoptosis in cerebral ischemic stroke. CNS Neurosci Ther 2013;19:813–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Q, Qian Z, Pan L, et al. Hypoxia-inducible factor 1 mediates the anti-apoptosis of berberine in neurons during hypoxia/ischemia. Acta Physiol Hung 2012;99:311–23 [DOI] [PubMed] [Google Scholar]
- 50. Cheng YL, Park JS, Manzanero S, et al. Evidence that collaboration between HIF-1alpha and Notch-1 promotes neuronal cell death in ischemic stroke. Neurobiol Dis 2014;62:286–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. LeComte MD, Shimada IS, Sherwin C, et al. Notch1-STAT3-ETBR signaling axis controls reactive astrocyte proliferation after brain injury. Proc Natl Acad Sci USA 2015;112:8726–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu Q, Fan X, Zhu J, et al. Co-culturing improves the OGD-injured neuron repairing and NSCs differentiation via Notch pathway activation. Neurosci Lett 2014;559:1–6 [DOI] [PubMed] [Google Scholar]
- 53. Sun F, Mao X, Xie L, et al. Notch1 signaling modulates neuronal progenitor activity in the subventricular zone in response to aging and focal ischemia. Aging Cell 2013;12:978–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao J, Sui M, Lu X, et al. Electroacupuncture promotes neural stem cell proliferation and neurogenesis in the dentate gyrus of rats following stroke via upregulation of Notch1 expression. Mol Med Rep 2015;12:6911–7 [DOI] [PubMed] [Google Scholar]
- 55. Muchnik C, Olivar N, Dalmasso MC, et al. Identification of PSEN2 mutation p. N141I in Argentine pedigrees with early-onset familial Alzheimer's disease. Neurobiol Aging 2015;36:2674–7.e1 [DOI] [PubMed] [Google Scholar]
- 56. Shi Z, Wang Y, Liu S, et al. Clinical and neuroimaging characterization of Chinese dementia patients with PSEN1 and PSEN2 mutations. Dement Geriatr Cogn Disord 2015;39:32–40 [DOI] [PubMed] [Google Scholar]
- 57. Tremolizzo L, Susani E, Mapelli C, et al. First report of PSEN2 mutation presenting as posterior cortical atrophy. Alzheimer Dis Assoc Disord 2015;29:249–51 [DOI] [PubMed] [Google Scholar]
- 58. Xia M, Chen S, Shi Y, et al. Probable novel PSEN2 Pro123Leu mutation in a Chinese Han family of Alzheimer's disease. Neurobiol Aging 2015;36:3334.e13–e18 [DOI] [PubMed] [Google Scholar]
- 59. Schroeter EH, Kisslinger JA, Kopan R.. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 1998;393:382–6 [DOI] [PubMed] [Google Scholar]