FIGURE 1.
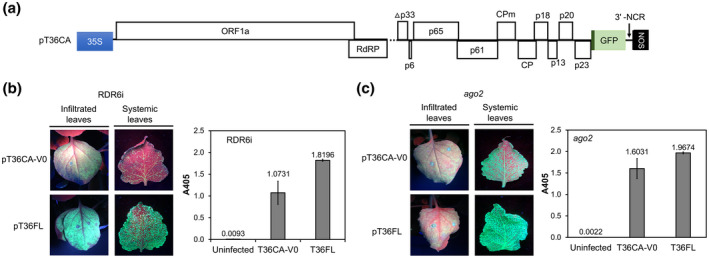
T36CA and its infectivity in RNA silencing‐defective Nicotiana benthamiana plants. Two prototype vectors, pT36CA‐ori and pT36CA‐V0, were constructed and inoculated to RDR6i and ago2 N. benthamiana plants by agroinfiltration and evaluated for bioactivity. (a) A schematic map of the T36CA binary plasmid (pT36CA) illustrating the region between the cauliflower mosaic virus 35S promoter and nopaline synthase (NOS) terminator. White boxes represent open reading frames (ORFs) encoding a polyprotein (ORF1a), the RNA‐dependent RNA polymerase (RdRP), the minor coat protein (CPm), the major coat protein (CP), and proteins named according to their predicted molecular weights (numerals preceded by the letter “p”). A gene encoding green fluorescent protein (GFP), expressed under the control of a controller element of T36CA CP (dark green), is inserted between p23 and the 3′ noncoding region (NCR). The dashed line represents the region in p33 deleted (Δp33) in pT36CA. (b) and (c) GFP fluorescence (left panels) was observed in the infiltrated (attached) and systemic (detached) leaves of N. benthamiana lines RDR6i (in the presence of exogenous HC‐Pro) (b) and ago2 (c) following the agroinfiltration of pT36CA‐V0 or pT36FL. Virus accumulation was determined by double antibody sandwich‐ELISA (right panels) at 9 weeks postinfiltration. The A405 value for each virus/treatment indicated on the x axis is the average for four individual samples from a representative experiment (with error bar representing SE)
