FIGURE 1.
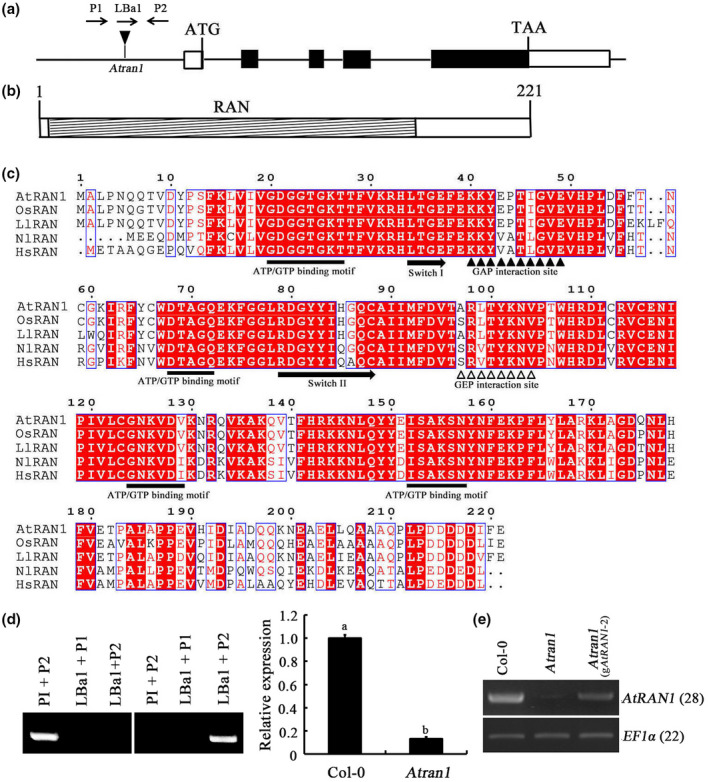
Structure of theAtRAN1gene and AtRAN1. (a) Structure ofAtRAN1. Black boxes, coding sequence; white boxes, 5′ (80 nt) and 3′ (347 nt) untranslated regions; horizontal line, intron sequence. (b) Structure of AtRAN1. Hatched box, RAN domain (amino acids 7–171). (c) Sequence alignment and structures of RANs fromArabidopsis(AED92779.1),Nilaparvata lugens(KT313028.1),Lepidium latifolium(GU014818.1),Homo sapiens(BC051908.2), andOryza sativa(AB015287.1). Filled rectangles, domain elements. Amino acids with 100% conservation are shaded red. Gaps were introduced to permit alignment. (d) PCR verification of theAtran1T‐DNA mutant and quantitative reverse transcription PCR (RT‐qPCR) analysis of the homozygous T‐DNA insertion line to determine the level ofAtRAN1transcript relative to Col‐0 plants.S16andEF1αwere used as internal controls. (e) Recovery ofAtRAN1expression in theAtran1mutants complemented with wild‐typeAtRAN1gene.AtRAN1transcript levels were determined by semiquantitative RT‐PCR
