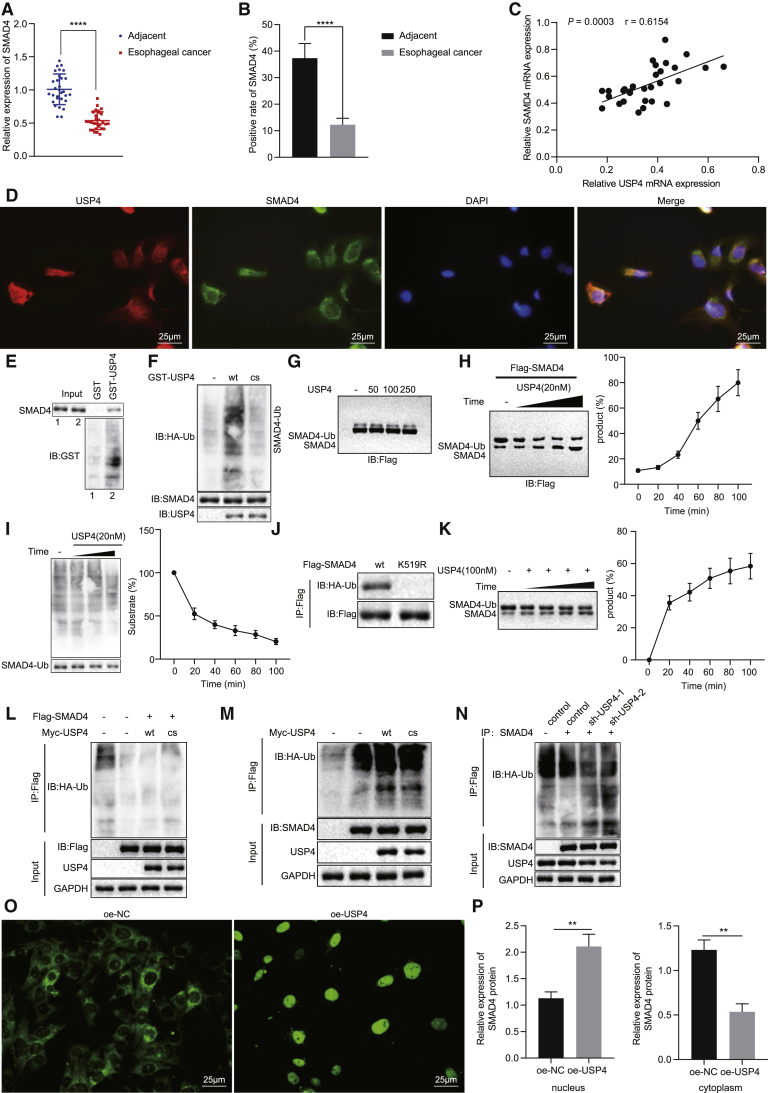
Figure 4.
USP4 Overexpression Increases the Deubiquitination and Translocation of SMAD4 into Nuclei in Cells
(A) SMAD4 mRNA expression in EC tissues and adjacent normal tissues detected by qRT-PCR (n = 30). (B) Quantitation of SMAD4 protein in EC tissues and adjacent normal tissues by immunohistochemical analysis (n = 30). (C) Pearson correlation analysis of USP4 expression with SMAD4 expression. (D) Immunofluorescence detection of SMAD4 and USP4 localization in TE-1 cells (×400). (E) GST pull-down detection of SMAD4 and USP4 interaction. (F) coIP detection of the effect of USP4 on SMAD4 ubiquitination. (G) Immunoblotting detection of ubiquitination of SMAD4 after 30-min co-incubation of different concentrations of USP4 with purified Flag-SMAD4. (H) Western blot analysis of the effect of the optimal concentration (20 nm) of recombinant USP4 on the deubiquitination of SMAD4. (I) The effect of the optimal concentration (20 nm) of recombinant USP4 on the deubiquitination of SMAD4. (J) Immunoblotting analysis of whole-cell lysate and immunoprecipitate of HEK293T cells with stable expression of HA-Ub after transfection of Flag SMAD4 or Flag-SMAD4 K519R. (K) Ubiquitination status of SMAD4 after incubation of 508-529 covalent ubiquitin peptide with 100 nM USP4 on K519 for different times. (L) SMAD4 ubiquitination status was observed by immunoblotting after affinity purification of the tagged SMAD4 protein in TE-1 cells. (M) Western blot analysis of ubiquitination of SMAD4 in cells stably expressing HA-Ub after transfection with Myc-USP4-WT or Myc-USP4-CS, respectively. (N) Western blot analysis of the effect of USP4 inhibition on the ubiquitination of SMAD4 in cells stably expressing HA-Ub. (O) Immunofluorescence detection of SMAD4 expression in cytoplasm and nucleus (×400). (P) Western blot analysis of SMAD4 protein in cytoplasm and nucleus of TE-1 cells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Paired t test was used for comparison between data of EC tissues and adjacent normal tissues, while unpaired data between the other two groups were compared by unpaired t test. Comparisons among multiple groups were performed using one-way ANOVA. Data are shown as mean ± standard deviation of three technical replicates.