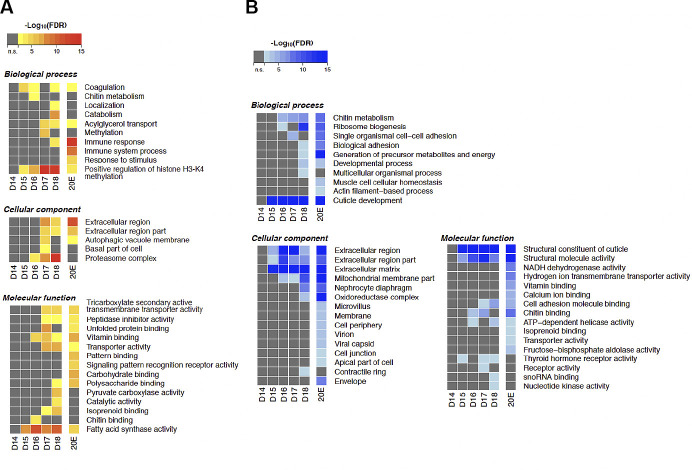
Fig. 3.
Gene Ontology (GO) enrichment analysis of differentially expressed genes during intersegmental muscle (ISM) development. A: GO terms enriched with the upregulated genes between day 13 and the other time points during the ISM development. The color shows the statistical significance measured in false discovery rate (FDR). B: GO terms enriched with the downregulated genes between day 13 and the other time points during the ISM development. The color shows the statistical significance measured in false discovery rate. n = 3 independent libraries were analyzed for each developmental stage. Atrophy (days 15–17) was associated with increased coagulation, histone H3-K4 methylation, acylglycerol transport, autophagy, fatty acid synthase activity, and vitamin binding and the repression of chitin metabolism, adhesion, and ATP-dependent helicase activity. The commitment to die on day 18 involved enhanced protein catabolism, immune response, the proteasome, membrane transporters, fatty acid synthase, and DNA methylation and the repression of ribosome production, cuticle formation, cell adhesion, and nucleotide kinase activity. 20-Hydroxyecdysone (20E) prevented most of the changes in death associated processes. D, day.