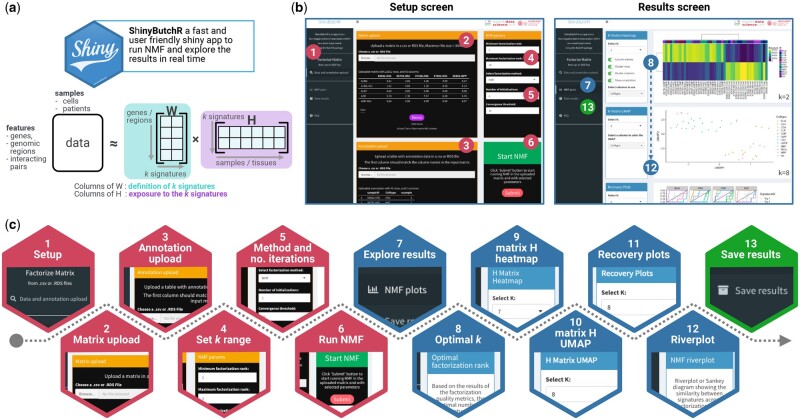
Figure 1.
schematic representation of a ShinyButchR NMF-based workflow. (a) ShinyButchR takes a non-negative matrix as input to perform NMF, decomposing the input matrix into a signature matrix W and an exposure matrix H. (b) Main screens of ShinyButchR user interface. The panel on the left shows the “Setup screen” of the app, where the user can upload a dataset and associated annotation table, as well as tuning the parameters to run the matrix decomposition. The panel on the right shows the “Results screen”, where the user can explore the results interactively, e.g., selection of the optimal factorization rank, clustering analysis, association to known biological and clinical factors and signature stability assessment. (c) Steps performed in the ShinyButchR workflow, the setup steps (i.e., steps 1 to 6) are shown in red, the results exploration steps (i.e., steps 7–12) are shown in blue, and the final save results step (i.e., step 13) is shown in green.