Abstract
Rapid and widespread changes in brain anatomy and physiology in the first five years of life present substantial challenges for developmental structural, functional, and diffusion MRI studies. One persistent challenge is that methods best suited to earlier developmental stages are suboptimal for later stages, which engenders a trade-off between using different, but age-appropriate, methods for different developmental stages or identical methods across stages. Both options have potential benefits, but also biases, as pipelines for each developmental stage can be matched on methods or the age-appropriateness of methods, but not both. This review describes the data acquisition, processing, and analysis challenges that introduce these potential biases and attempts to elucidate decisions and make recommendations that would optimize developmental comparisons.
Keywords: Development, Neuroimaging, Infant, Child, Brain, MRI
1. Introduction
Early childhood is characterized by rapid brain development (Gilmore et al., 2018) and represents a critical time for establishing developmental trajectories of neural anatomy and physiology. Consequently, experiences are most influential during these early years, with the potential to shape and alter typical developmental trajectories (Nelson and Gabard-Durnam, 2020). Magnetic resonance imaging (MRI) technology, which began as a tool to study disorders and aberrations in adult clinical populations, was brought to bear on typical infant and early childhood brain structure (Pfefferbaum et al., 1994; Raininko et al., 1994) and function (Born et al. 1998, 2000; Martin et al., 1999) over two decades ago. With the ability to study the brain at early stages of life came the opportunity to understand developmental changes and trajectories using a longitudinal time frame. However, substantial and rapid changes in brain anatomy and physiology present significant and unique challenges for data acquisition, processing, analysis, and interpretation as part of a developmental neuroimaging pipeline.
Understanding neurodevelopmental changes across early childhood requires accurate and precise measurements of brain structure and function at all developmental stages (or age groups), and in a manner that does not favor a particular developmental stage when they are compared. On the one hand, past critiques have highlighted general methodological concerns for developmental studies (Madhyastha et al., 2018; Vijayakumar et al., 2018; or the "Methodological Challenges in Developmental Neuroimaging: Contemporary Approaches and Solutions" Issue (Vol. 33, 2018) of Developmental Cognitive Neuroscience journal). However, these do not address the neurodevelopmental processes unique to children under five years of age, which are qualitatively and quantitatively different and often considerably faster than those transpiring after year five. On the other hand, critiques specifically for infant studies do not address developmental comparisons (Mongerson et al., 2017; Makropoulos et al., 2018; Zhang et al., 2019; Dubois et al., 2020). Indeed, there are methodological challenges that are unique to studies on early development that warrant special consideration (Vasung et al., 2019). In the present article, we first outline macroscopic anatomical and physiological changes that can impact developmental neuroimaging studies of early childhood. Grounded in these developmental changes, we then review the unique challenges associated with conducting developmental MRI studies in infants (i.e., children ≤ 1 year) and young children (1−5 years) and highlight various approaches that have been developed to overcome these challenges. We conclude by outlining where the field continues to struggle and what future methodological developments are needed. Overall, the aim of this review is to provide a resource for trainees and junior investigators, as well as for established investigators who are starting to conduct MRI studies in this age range.
2. Developmental changes in brain anatomy and physiology
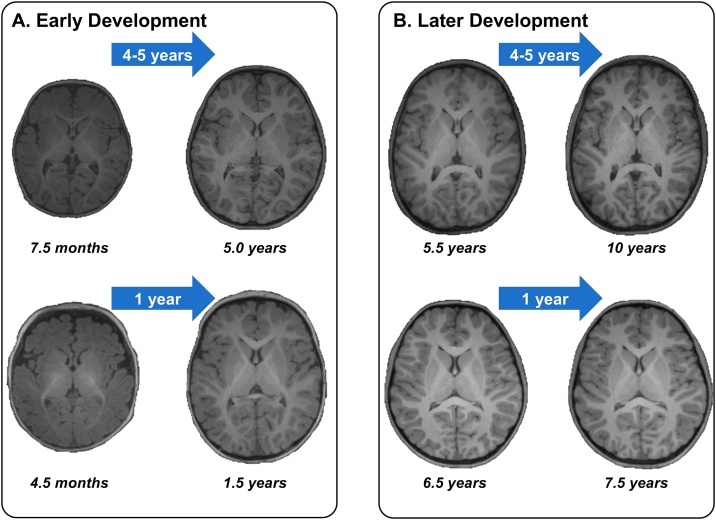
Proportionally, the largest neuroanatomical growth transpires in the first year of life and tapers thereafter (Matsuzawa et al., 2001; Knickmeyer et al., 2008; Groeschel et al., 2010; Sanchez et al., 2011; Deoni et al., 2012; Geng et al., 2012; Gilmore et al., 2012; Li et al., 2013; Lyall et al., 2015). Namely, total intracranial volume, cortical gray matter volume, subcortical volume, and cerebellar volume more than double in the first year (Knickmeyer et al., 2008; Gilmore et al., 2012). A smaller, yet significant, increase is observed in cortical thickness (Lyall et al., 2015), gray matter surface area (Li et al., 2013; Lyall et al., 2015), white matter volume (Knickmeyer et al., 2008), and white matter fractional anisotropy (FA; Geng et al., 2012), a measure of the structural organization of white matter pathways. The relative growth for the majority of these brain measures diminishes substantially in year two (Knickmeyer et al., 2008; Geng et al., 2012; Gilmore et al., 2012; Li et al., 2013; Lyall et al., 2015), and afterward for gray and white matter (Courchesne et al., 2000; Matsuzawa et al., 2001; Groeschel et al., 2010) and FA (Reynolds et al. 2019). In contrast, relative growth in cortical white matter increases (Knickmeyer et al., 2008). Importantly, these growth rates also differ by brain region (Gilmore et al., 2012) and white matter tract (Deoni et al., 2012; Geng et al., 2012). Please see Fig. 1 for sample MRI images depicting relatively rapid brain growth in early compared with later development. However, comprehensive examinations of brain anatomy are currently limited to the first two years following birth (Table 1). Further work is needed to extend developmental trajectories to five years.
Fig. 1.
Sample structural MRI images depicting brain growth during (A) early and (B) later development. Each quandrant shows brain images from a child at two developmental stages as collected longitudinally. Please note the substantial anatomical changes during early development, especially in the first year of life, compared to later development.
Table 1.
Anatomical growth estimates for brain and skull.
| Anatomical Growth | Year 1 | Year 2 | Year 3 |
|---|---|---|---|
| Total Intracranial Volumea | 101 % | 15 % | |
| Cortical Gray Matter Volumea, b | 129 % | 17 % | |
| Cortical White Matter Volumea | 11 % | 19 % | |
| Subcortical Volumea, b, c | 117 % | 14 % | |
| Cerebellar Volumea | 240 % | 15 % | |
| Cortical Surface | 78 % | 20 % | |
| Cortical Thickness | 31 % | 4.3 % | |
| Fractional Anisotropy | 30 % | 7.8 % | |
| Head Circumference | 30 % | 5.2 % | 2.3 % |
| Skull Thickness | 38 % | 28 % | 22 % |
Values summarized from Knickmeyer et al., 2008; Geng et al., 2012; Gilmore et al., 2012; Li et al., 2013, 2015a,2015b; Lyall et al., 2015, Centers for Disease Control and Prevention, National Center for Health Statistics).
Average of min and max range of Knickmeyer et al. (2008).
Average of Knickmeyer et al. (2008) and Gilmore et al. (2012).
Includes Brainstem for Knickmeyer et al. (2008).
In addition to developmental changes in brain tissue, anatomical structures that support the brain also change. For instance, head circumference increases most in the first year of life, while this increase is significant but smaller in the second and third years of life (Centers for and Disease Control and Prevention, 2001). Average skull thickness also increases most in year one, although it still increases substantially in years two and three (Table 1;Li et al., 2015a). Meanwhile, angiogenesis (i.e., the growth and branching of blood vessels) vastly expands microvasculature established prenatally; e.g., the dense layer of veins in vascular layer 3 is pruned and supplanted by a plexus of capillaries (Norman and O’Kusky, 1986), which scales with local metabolic activity and neural and glial (e.g., astrocyte) demands for oxygen. Considerable microvascular remodeling also occurs during this time, involving vessel sprouting, pruning, and lengthening (Harb et al., 2013; for a review of postnatal microvascular development in the brain, please see Coelho-Santos and Shih, 2019). Downstream, developmental changes in vasculature can affect functional MRI and near infrared spectroscopy experimentation (for a review, please see Vasung et al. (2019)), as these measures derive from neurovascular and gliovascular interactions; i.e., neurons and glial cells consume and deplete oxygen to function and this oxygen is replenished by oxygenated hemoglobin via blood vessels (Yu et al., 2010). Finally, heart rate, respiration, and water content all decrease between infancy and adulthood (Mongerson et al., 2017; Bastiani et al., 2019).
These anatomical and physiological changes have profound implications for conducting structural and functional developmental MRI studies for two main reasons. First, methods that were developed for adults and older children need to be adapted to the developmental stage of the infant or young child to optimize the accuracy and precision of brain measures (e.g., tissue volume as a measure of brain structure, or functional activation and functional connectivity as measures of brain function). Second, practices that are optimal for a child of one age may be suboptimal for a child of a different age and induce spurious, age-related differences that are driven by methodological inconsistencies, rather than true brain development. Ultimately, this leads to a trade-off and challenging decision for investigators of whether to optimize methods for the neuroanatomy and neurophysiology unique to the specific developmental stage, or preserve consistent methods across stages. In the case of the former, developmental stages are matched on whether methods are optimal for children of that age; in the case of the latter, comparisons of developmental stages are matched on the methods themselves. Both options have potential benefits, but also introduce biases.
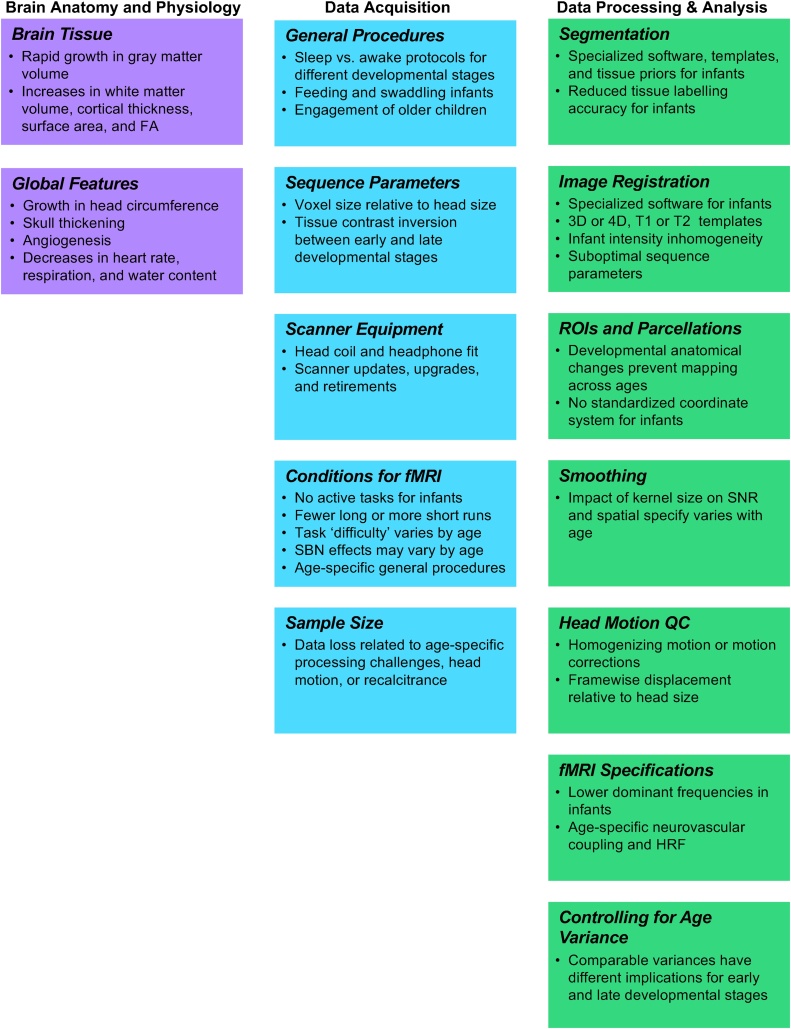
In the following sections, we highlight and describe the acquisition, processing, and analysis steps involved in developmental MRI datasets that may introduce these biases (please see Fig. 2 for a summary). To ease implementation for any trainees and investigators entering the field, we have structured these sections largely according to an experiment pipeline typical of an early developmental MRI study.
Fig. 2.
Factors that may introduce bias when scanning children at different developmental stages between birth and age five.
3. Data acquisition
Acquiring data for developmental MRI studies in children ages 0−5 years presents numerous challenges. The first set of challenges relates to the procedures for scanning young children. Infants and toddlers require specialized procedures and the absence of these can hamper the quality of data acquisition (Raschle et al., 2012a). Second, sequence parameters and scanner equipment that are optimal for infants may differ from those that are optimal for children between one and five years, given the current understanding of widespread and rapid developmental changes in brain and head anatomy and physiology. Third, angiogenesis and other developmental changes that affect neurovasculature have profound implications for acquiring functional MRI (fMRI) data, the signal for which depends on neurovascular coupling (Vasung et al., 2019). Each step in the data acquisition pipeline carries with it a trade-off between implementing age-appropriate data acquisition practices or using consistent methods across developmental stages. Lastly, as data processing is particularly challenging for children 0−5 years, data loss is likely to be high. This should be considered when estimating sample sizes for developmental stages in this age range.
3.1. General procedures
Procedures for pediatric imaging require careful consideration to maximize compliance and data quality. One reason that procedures for MRI data acquisition in developmental neuroimaging for ages 0−5 are challenging is because scanning infants and toddlers requires adapted data acquisition procedures (Raschle et al., 2012a). One example of an age-specific procedure relates to whether the child is asleep or awake during the scan. For the youngest children (i.e., birth to three years old), a natural sleep protocol is often used because children of this age can sleep in the scanner (Liu et al., 2008; Fransson et al., 2009; Gao et al., 2009; Doria et al., 2010; Graham et al., 2016; Howell et al., 2019; Turesky et al., 2019, 2020, Yin et al. (2020)), which reduces motion and increases the likelihood of collecting usable data. Common techniques include a form of “feed and wrap,” in which infants are fed prior to the scan and swaddled during the scan (Almli et al., 2007; Ibrahim et al., 2015), while more involved techniques can rely on vacuum immobilization (Dean et al., 2014; Hughes et al., 2017; for techniques used to induce natural sleep in infants during MRI, please see Raschle et al., 2012a; Antonov et al., 2017). Notably, this natural sleep procedure is in general most successful in the youngest children, although this is subject to large inter-individual variability. Infants have also been scanned while awake (Deen et al., 2017), more recently during cognitive tasks (Ellis et al., 2020), though scanning awake infants is rarer due to higher likelihood of motion. For typical developing children closer to five years of age, the natural sleep protocol is used less often and supplanted by detailed protocols that are specifically designed to engage the child, such as watching movies during structural sequences or rebranding functional tasks as games (Raschle et al., 2011, 2012; Vanderauwera et al., 2017; Yu et al., 2018). Some developmental studies from birth to age five have been performed with all children asleep in the scanner (Gabard-Durnam et al., 2018), while others allow older children (≥ 3 years) to remain awake (depending on caregiver preference) and younger children (< 3 years) to sleep (Howell et al., 2019), which can introduce bias.
Moreover, this age and sleep/awake-related bias may be compounded when examining certain clinical populations, who tend to exhibit greater motion, such as children with attention-deficit/hyperactivity disorder or autism spectrum disorder. Taken together, mitigating head motion across early development may require different procedures prior to and during data acquisition. However, as discussed in greater detail below, this practice also introduces confounds for functional neuroimaging as sleep state and sleep stage can affect these measures (Horovitz et al., 2009; Picchioni et al., 2013).
3.2. Sequence parameters
Sequence parameters are the set of radiofrequency pulse and gradient properties that the MRI scanner uses to acquire images, which vary considerably depending on whether one wishes to acquire structural, functional, or diffusion images and also on the population examined (Saunders et al., 2007). MRI scanners acquire images at the level of the voxel, a three-dimensional unit in the shape of a cube or rectangular prism. Analogous to pixels on a computer screen, each voxel occupies a small fraction of a larger three-dimensional grid or volume and contains a corresponding intensity. The voxel size, which dictates the resolution of an image, can be set through the following sequence parameters: field-of-view, matrix size (i.e., number of samples in each direction), and slice thickness. Meanwhile, the signal-to-noise ratio (SNR) of an image, which reflects desired signal intensity compared with the level of background noise (Edelstein et al., 1986), can also be affected by these sequence parameters (Redpath, 1998), as voxel size and SNR are directly proportional (Runge et al., 2013). Resolution and SNR are critical image features because—along with tissue contrast, scan time, and presence of artifacts—they directly contribute to overall image quality. Accordingly, sequence parameters for developmental MRI studies need to be set such that voxel sizes and intensities are acquired without bias toward one or more developmental stage. This proves difficult when considering that the optimal set of sequence parameters (e.g., to maximize resolution and SNR) for one developmental stage may be suboptimal for another. Therefore, broadly, a second set of challenges for early developmental neuroimaging relates to whether to employ age-appropriate sequence parameters that optimize image quality for each developmental stage or identical sequence parameters for all stages.
This trade-off is most salient when considering the substantially smaller brains of infants by comparison to those of older children. This has important implications when considering which voxel sizes to employ and whether to optimize for each developmental stage or standardize across stages. First, the same voxel size used for younger and older children will represent a much larger proportion of brain tissue at the earlier developmental stages, which biases the resolution in favor of older children and in a manner that cannot be overcome through normalization or smoothing (please see Data processing and analysis section below). Furthermore, the substantial growth of individual brain structures across early development (Gilmore et al., 2012) means that fewer voxels will capture each brain structure in the earlier compared with later developmental stages (Keil et al., 2011). Second, due to the smaller brain structures in younger compared with older children (Gilmore et al., 2012), single voxels are more likely to include multiple brain structures and tissue types, reducing the accuracy of segmentation of tissue types (discussed in greater detail below) and brain structures. And third, the smaller head size in children can cause a substantial drop in contrast between tissues compared with adults (Prastawa et al., 2005).
Rapid developmental changes in brain anatomy and physiology also cause brain tissue imaged with the same MRI sequence to look vastly different for children of different developmental stages (Fig. 1). For instance, T1-weighted sequences, which are commonly employed in MRI studies for their superior tissue contrast and clear visualization of brain structures, show white matter as brighter (i.e., higher intensity) than gray matter in adults and children over age one (Gilmore et al., 2018). By comparison, prior to six months, with increasing myelination (Deoni et al., 2012) and decreasing water content (Counsell and Rutherford, 2002), gray matter exhibits higher intensity than white matter on T1-weighted sequences; gray and white matter exhibit more comparable intensity between ages 8–12 months (Paus et al., 2001). While these differences may meaningfully characterize developmental changes in brain tissue, they also cause challenges for adequately measuring this brain tissue (e.g., in terms of volume or thickness). For instance, contrast between gray and white matter increases across developmental stages and can lead to better tissue labelling in older compared to younger children (also, please see Segmentation subsection below). Thus, if the aim were to produce images with corresponding relative tissue intensities, then different developmental stages would require different scan sequences. For structural scans, children over one year would undergo T1-weighted sequences and infants under one year would be scanned with T2-weighted sequences (Paus et al., 2001; Gilmore et al., 2018), which rely on different types of proton relaxations (after protons are perturbed by radiofrequency pulses). In addition, intensity inhomogeneity, in general, caused by heterogeneous static and radiofrequency fields, becomes more problematic in infants when considering ongoing myelination, as well as higher variability in tissue intensity and low-intensity contrast (Prastawa et al., 2005).
Overall, when deciding on sequence parameters, developmental neuroimaging studies encounter a central trade-off between optimizing these parameters for the neuroanatomy and neurophysiology unique to specific developmental stages, or using the same parameters across stages. Both approaches introduce benefits and biases. The first option, optimizing sequence parameters to a specific developmental stage, can increase the quality of images acquired for children of that age and improve the accuracy with which brain measures are estimated. By comparison, using suboptimal parameters could reduce image quality, which in turn could lead to overestimation of volumetric, surface, and thickness estimates of one tissue type and underestimation of the other (relative to manual estimations). In this sense, tailoring sequences for each developmental stage, to more accurately estimate brain measures, can reduce bias in developmental comparisons. The second option, preserving scanning parameters across stages, is more common in developmental research. This approach may increase comparability across age ranges because no differences can be attributed to differences in sequence parameters, but it may also introduce bias from reduced image quality in specific developmental stages that are scanned using parameters that are not optimized for their unique biology. It is possible that the bias introduced by suboptimal parameters is more severe for developmental studies of early childhood and has been hitherto underestimated when considering the early childhood population; further research will be needed to determine whether this is the case.
3.3. Scanner equipment
In addition to scanning procedures and acquisition parameters, investigators face challenges in choosing MRI scanner equipment for studying early development. For instance, head coils designed to optimize infant imaging are too small for older children. Although head coils designed for older children can accommodate a smaller infant head, the SNR, encoding performances, and consequent image quality will all be lower compared to when using an age-appropriate head coil because the gap between the head and head coil is wider (Keil et al., 2011). In the last decade, several laboratories and vendors have developed equipment that optimizes pediatric MR imaging for specific ages; however, the use of age-specific head coils introduces other challenges. First, they can be difficult to obtain, as they are often custom-built and costly (Keil et al., 2011). Second, if the head coil used for one developmental stage yields higher SNR compared with the head coil for another stage, then confounds are introduced. And third, even if SNR is comparable, the equipment used for each group is not consistent, which forces a trade-off between standardizing the head coil or the fit of the head coil across stages. Importantly, if investigators attempt to employ sequences with smaller voxel sizes to capture the smaller structures in infants (as described in the previous section), then SNR and consequent image quality would inherently decrease. The improvements in SNR from an age-appropriate head coil could offset the loss in SNR from the reduced voxel size. Additionally, age-appropriate head coils are also likely to improve comfort for children (Keil et al., 2011), which can increase time tolerated inside the scanner while also reducing head motion. As for head coils, investigators may also need multiple headphones for functional MRI studies to accommodate differing head sizes.
If conducting longitudinal studies, then scanner updates, upgrades, and retirements also pose challenges for conducting multiple longitudinal measures of one individual. For example, infants may undergo MRI with one scanner; but by the time they are five years old, that scanner may have gone through several upgrades. Or perhaps the institution conducting the study replaces the scanner used for the infants with a newer scanner with superior image quality. As the field moves toward using larger datasets, one scanner at one site may be insufficient to acquire all of the scans planned for a developmental study. The use of different scanners at different scanning sites can alleviate this challenge, but doing so might also bias comparisons between developmental stages if each site scans only children of particular ages. Future work needs to assess whether current guidelines for conducting multi-site studies (e.g., Glover et al., 2012) and practices for integrating datasets acquired from multiple sites or scanners (i.e., harmonization; Karayumak et al., 2019; Pomponio et al., 2020) can be applied to early developmental studies without introducing additional confounds.
3.4. Conditions for fMRI
fMRI captures neural responses by acquiring multiple volumes while participants perform a task or rest (often awake and mind-wandering, or asleep). Experimental conditions need to be designed to enable participant compliance that is consistent across developmental stages. Meeting this goal is especially difficult when examining developmental changes between birth and age five, foremost because infants can perform only passive tasks such as processing auditory or visual stimuli (e.g., faces, bodies, and scenes) while awake (Deen et al., 2017) or, while asleep, viewing flickering lights through closed eyelids (Born et al., 1998) or listening to sounds (Anderson et al., 2001). Other studies have acquired fMRI data while children were asleep without imposing a task (i.e., a resting-state fMRI paradigm; Fransson et al., 2007, 2011; Doria et al., 2010; Gao et al., 2015a, 2015b;Graham et al., 2015a, 2015b; Pruett et al., 2015; Graham et al., 2016; Mitra et al., 2017; Rudolph et al., 2018; Turesky et al., 2019). In theory, passive task and resting-state paradigms can also be used for older children, making them of interest for developmental studies. In practice, however, as children emerge from early infancy, they do not require as much sleep and have more difficulty remaining motionless in the scanner during passive tasks and resting-state, introducing greater motion into the older compared with earlier developmental stages. For task paradigms, fewer long functional runs could mitigate motion in infants and toddlers because these offer a consistent environment in which children can sleep without interruption. However, for older children, dividing a long functional run into multiple shorter functional runs reduces the amount of time the child needs to remain motionless and still facilitate collection of a sufficient number of data points; e.g., our group has probed phonological processing in children using separate experimental and control conditions (Yu et al., 2018). In contrast, resting-state paradigms, which are mainly used to examine functional connectivity, require at least four minutes of continuous data acquisition (after any data removal due to scrubbing; Power et al., 2012) for functional connectivity estimates to stabilize, according to studies in adolescents (Satterthwaite et al., 2013) and young adults (Van Dijk et al., 2010). If a comparable minimum duration is needed for infants (Mongerson et al., 2017), then separating this paradigm into multiple runs and concatenating volumes cannot be a solution. Lastly, even if children are given identical tasks across stages, task difficulty can vary inversely by age, introducing another source of bias: in infants, the task may be too difficult, but then too easy by age five, .
Additionally, most fMRI experiments rely on the blood-oxygen-level-dependent (BOLD) signal to measure brain activation. The BOLD signal, which reflects the ratio of oxygenated to deoxygenated hemoglobin, is affected by cerebral blood flow, cerebral blood volume, and cerebral metabolic rate of oxygen (Ogawa and Lee, 1990; Raichle, 1998). Therefore, angiogenesis and microvascular refinement in early development could directly affect the BOLD signal (please see Developmental changes of brain anatomy and physiology section and Coelho-Santos and Shih (2019) and Vasung et al. (2019) for reviews).
The BOLD signal can also be affected by other, distal factors that may differ by developmental stage. For instance, as described above, infant scans are mainly conducted while infants are asleep, whereas older children are not generally instructed to sleep. If children of earlier developmental stages are asleep and later developmental stages are awake, then resulting developmental effects could instead reflect differences in consciousness, as these—with differential influences on cerebral metabolic processes—affect the BOLD signal and subsequent functional measures differently (Horovitz et al., 2009; Picchioni et al., 2013). Evincing this, one report has shown that resting-state functional connectivity in sleeping infants is qualitatively more similar to resting-state functional connectivity in adults during sleep relative to adults during wakefulness (Mitra et al., 2017). In addition, identical patterns of resting-state functional connectivity do not underlie all sleep stages. This variability may be monitored using concurrent EEG or by tracking physiological measures such as heart rate or respiration. However, EEG acquisition in developmnental populations has its own set of challenges, especially when performed concurrently with MRI.
Another factor known to impact consciousness during acquisition pertains to scanner background noise (SBN), which can be extremely loud at over 100 decibels. SBN can interfere with sleep, which can negatively impact head motion, but it also requires special consideration in the context of fMRI as it has been shown to attenuate auditory cortex responses (Gaab et al., 2007a, 2007b, 2008) and these effects may vary by age. Ear protection has effectively attenuated SBN in younger children (Zahr and de Traversay, 1995). However, keeping ear protection fastened can be more challenging in infants and toddlers compared with older children. Furthermore, it is important to consider that often only one set of headphones are available at an imaging center and their fit may differ depending on the age of the child. Certain experimental designs, such as sparse sampling (whereby auditory stimuli are played during silent delays in MRI volume acquisitions; Gaab et al., 2003; Perrachione and Ghosh, 2013), which reduces effects of SBN (Gaab et al., 2007b), can be alternatives. Therefore, investigators should consider whether these adjustments can be made to their data acquisition protocols in a manner that does not limit their ability to address their research questions. However, these sparse protocols increase overall scanning duration and contain long periods of silence, which can be suboptimal for certain age ranges as it can lead to increased head movement. Therefore, investigators should consider whether these adjustments can be made to their data acquisition protocols in a manner that does not limit their ability to address their research questions.
Lastly, systematic differences in internal and external conditions among developmental stages prior to and throughout data acquisition may bias fMRI data acquisition. For instance, infant feeding as part of the “feed and wrap” technique (Almli et al., 2007; Ibrahim et al., 2015) could affect glucose metabolism, which would affect the BOLD signal. Also, anxiety and discomfort evoked by scanner background noise (Gaab et al., 2007a, 2007b;Raschle et al., 2012a) may disproportionally affect infants’ and older children’s responses to stimuli. Differences in scan state (environment) too may bias fMRI data acquisition as functional brain networks constituting functionally coupled brain regions (Buckner et al., 2013) exhibit moderate variability depending upon whether data are acquired while the participant is performing a task or at rest (Cole et al., 2014; Gratton et al., 2018). For example, with infants swaddled (Almli et al., 2007) or vacuum immobilized (Dean et al., 2014; Hughes et al., 2017) to induce sleep, and older children physically unconstrained, somatosensory inputs would differ by developmental stage. Overall, different experimental conditions for different developmental stages may introduce bias during fMRI data acquisition.
3.5. Sample size
The challenges described above have implications for proposed sample sizes, attrition rates, and power calculations. First, infants may have difficulty sleeping (or remaining asleep) in the scanner, which could cause severe and repeated motion artifacts and require that they be removed from the scanner. Second, data processing challenges unique to the infant stage (e.g., segmentation with inverted and reduced gray/white matter contrast; please see below) could reduce the number of datasets of sufficient quality to be included in developmental analyses. In longitudinal studies, these challenges can disqualify participants from follow-up scans at later developmental stages if laboratories do not choose to impute in their statistical analyses. And third, head motion in older children who now may be awake in the scanner (please see General procedures subsection above) may reduce the number of usable datasets at later stages, particularly stymieing longitudinal analyses.
Concordantly, at roughly 70 % (e.g., Almli et al.), success rates for developmental studies from birth to five years are fairly low, and these rates vary by developmental stage. For instance, success rates tend to follow a U-shape across development, decreasing from birth to toddlerhood and then increasing closer to age 5. Specifically, children scanned at 2–4 weeks, 1 year, and 2 years had success rates of 60 %, 50 %, and 50 % respectively (Knickmeyer et al., 2008); and between 2 and 5 years, higher rates have been reported at 72 % (Thieba et al., 2018). A study by our group reported on this trend using cross-sectional data, showing 63 % success rates at 3–7 months, 59 % at 8–15 months, and 97 % at 4–6 years (Raschle et al., 2012a, 2012b). Success rates also vary depending upon the definition of success. For example, Thieba and colleagues (2018) reported a 72 % success rate for acquiring one out of three scans, but a 48 % success rate for acquiring all three scans (Thieba et al., 2018).
For longitudinal studies, investigators should also expect attrition and consequently success rates lower than those cited above. An attrition rate of approximately 20 % has been reported, though this figure can vary widely depending upon age, socioeconomic status, and psychiatric factors (Stange et al., 2018), as well as the duration of time between data acquisitions. To mitigate this, maintaining strong relationships with parents of children ages 0−5 years is extremely important – though uniquely challenging as this period of their lives is often accompanied with considerable flux, especially for those who have only recently become parents for the first time (please see Stange et al. (2018) for longitudinal neuroimaging retention practices and Teague et al. (2018) for general longitudinal retention practices). Guidelines in the field that resolve or mitigate these challenges will help to accurately estimate proposed sample sizes and attrition. However, in the interim, it may behoove investigators to begin with a higher initial sample size for developmental studies from birth to age five relative to the initial sample size used in developmental studies in children over five years of age.
4. Data processing and analysis
To quantitatively examine developmental stages, MRI data need to undergo rigorous pre- and post-processing. Data processing and analysis can comprise many steps, but we will limit our discussion to steps that are most likely to introduce bias to developmental comparisons. These are segmentation, image registration, defining regions-of-interests and parcellations, smoothing, head motion quality control (QC), and functional MRI (fMRI)specification. As with data acquisition, each processing step introduces a trade-off between implementing age-appropriate data acquisition practices or using similar or identical methods across developmental stages.
4.1. Segmentation
Basic tissue segmentation is the process of identifying and separating gray matter, white matter, and cerebrospinal fluid voxels, which can be important for quantifying tissue volumes, limiting subsequent analyses to certain tissue types, and/or registering images (Ashburner and Friston, 2005). Accurate tissue segmentation in an MR image, including extracting the brain from the skull (i.e., brain extraction or skull stripping), is considerably more challenging for infants compared with older ages (Makropoulos et al., 2018), which is a substantial concern as segmentation is the gateway to many subsequent analyses (Devi et al., 2015). Unmyelinated axons, ongoing myelination, low tissue contrasts (Prastawa et al., 2005), and thin cortex (Lyall et al., 2015) in infants relative to older children and adults have limited the accuracy with which canonical segmentation tools can label gray/white matter tissue in infants (Makropoulos et al., 2018).
These age-related, neurobiological characteristics have engendered two potential challenges for developmental MRI studies. First, segmenting brains of different ages requires different tools, techniques, and/or parameters. Several labs have developed segmentation tools specifically for infants, such as Infant Brain Extraction and Analysis Toolbox (iBEAT; Dai et al., 2013; version 2.0 has recently been released: https://ibeat.wildapricot.org/) and Infant FreeSurfer (Zöllei et al., 2020; https://surfer.nmr.mgh.harvard.edu/fswiki/infantFS; for a full list of neonate segmentation tools as of 2018, please see Makropoulos et al., 2018). In addition, investigators can label tissue manually, which is time consuming and labor intensive (Makropoulos et al., 2018), but can be an effective alternative to automated tools (de Macedo Rodrigues et al., 2015). With regard to techniques, segmentation algorithms employing age-appropriate tissue priors—typically gray matter, white matter, and cerebrospinal fluid probability maps in which voxel intensities reflect the likelihood that that voxel corresponds to a particular tissue class (Ashburner and Friston, 2005)—also reduce misclassification of tissue, compared with segmentations using adult tissue priors (Altaye et al., 2008). Tissue priors spanning multiple stages beginning at the perinatal period are now available as part of 4D templates (Oishi et al., 2018; please see below for details). Similarly, some investigators have opted for a 4D longitudinal approach (which, as the name suggests, can be applied in longitudinal studies), in which segmentations are performed on data from older children (for whom segmentations are generally more accurate due to improved tissue contrasts), and then used as tissue priors to inform segmentation in infants (Shi et al., 2011; Gilmore et al., 2012). Segmentation without tissue priors (Dhankhar et al., 2010; e.g., using Advanced Normalization Tools (ANTs; https://github.com/ANTsX/ANTs) Atropos with k-means) would theoretically circumvent this trade-off, but introduces other bias as the algorithms might then rely more heavily on tissue contrast, which is inherently poorer in younger children (Prastawa et al., 2005), especially between 8 and 12 months (Paus et al., 2001). For brain extraction protocols that rely on a template with skull (such as the ANTs antsBrainExtraction module), age-appropriate templates would also reflect developmental increases in head circumference and skull thickness (Centers for and Disease Control and Prevention, 2001; Li et al., 2015a). Importantly, laboratories have started to focus on the development of tools, templates, and tissue priors for segmentation as well as image registration and parcellation steps as outlined below (e.g., please see https://www.med.unc.edu/bric/ideagroup/software/).
Second, segmentation quality is age-dependent even with specialized tools and techniques. This can lead to under- or overestimation of tissue volumes, surface areas, and cortical thickness in the younger age group and therefore erroneous, age-related differences when compared to older age groups. Given that some toolkits (e.g., Statistical Parametric Mapping (SPM; https://www.fil.ion.ucl.ac.uk/spm/) 12) perform tissue labelling in concert with image registration as part of the unified segmentation framework (Ashburner and Friston, 2005), segmentation quality may also affect analyses of brain structure or function that require normalizing to standard space (please see below). As with data acquisition (please see above), segmentation procedures for developmental studies come with a careful trade-off between adapting approaches to the age-appropriate range or maintaining approaches for measurements across stages.
4.2. Image registration
Registration is the process of aligning images to one another, and different data processing steps may depend upon it for various reasons. For instance, to control for head motion throughout a functional run, functional volumes will be realigned (via x, y, and z translations and pitch, roll, yaw rotations) such that brain images in every volume overlap. To control for one individual’s head movement between scans of different modalities (e.g., structural and functional volumes), the image alignment procedure is often referred to as co-registration. In the special case of normalization, all individual brain images are aligned or spatially normalized to a standard brain image (i.e., a template), which is important for group analyses to ensure that the location of any given brain area in one participant spatially corresponds to the location of the same brain area in another participant.
Normalization can be especially challenging for developmental studies for two main reasons. First, the process of registering very young brains to templates (either through linear or non-linear procedures) can require specialized tools. For instance, the first version of iBEAT software relied on Hierarchical Attribute Matching Mechanism for Elastic Registration (Shen and Davatzikos, 2002) for registering individual and standard images (Dai et al., 2013). In our experience, popular MRI data processing software packages such as SPM or the voxel-based morphometry toolkit implemented through it have suboptimally normalized individual infant images. That these tools couple normalization with segmentation is consistent with the limited adaptability of unified segmentation algorithms (Beare et al., 2016). In contrast, the ANTs algorithms could register individual infant and template images (Turesky et al., 2019) and as ANTs was not designed specifically for younger children, it can also process data effectively for older children, precluding bias due to different tools (we have made our code from a relevant study available on github: https://github.com/TeddyTuresky/Resting-StateInfantsBangladesh2019). Crucially, accurate normalization (and segmentation) also depend upon rigorous correction for intensity inhomogeneity (Mongerson et al., 2017; Makropoulos et al., 2018), which is substantial in infants (Prastawa et al., 2005; please see Data acquisition: Sequence Parameters subsection). N4ITK, an intensity inhomogeneity correction module implemented through ANTs (Tustison et al., 2010), is commonly used for infants (Wu and Avants, 2012; Makropoulos et al., 2014; Turesky et al., 2019), but it is unclear whether optimal corrections for all developmental stages can be achieved by an identical set of parameters.
It is also unclear which templates are most appropriate for processing early developmental data. For infants, most groups use age-corresponding templates (Gao et al., 2015a, 2015b; Turesky et al., 2019), and often with secondary transformations to or from adult templates to draw on stereotaxic standards (Graham et al., 2015a, 2015b; Graham et al., 2016). On the other side of the early developmental age range, at five years, adult templates have been used (Raschle et al., 2012a, 2012b; Raschle et al., 2014). However, brain structure from birth to age five changes so rapidly (Matsuzawa et al., 2001; Groeschel et al., 2010; Sanchez et al., 2011) that using an adult template or even a template corresponding to the mean or median of the developmental age range investigated can potentially favor registration accuracy for some developmental stages over others, as use of age-inappropriate templates can lead to misclassifications of tissue and brain regions (Wilke et al., 2003).
Use of age-appropriate templates (i.e., a 4D template with one target image per age group) can reduce this confound (Oishi et al., 2018), though there is still the concern that normalization in the youngest children would not be as precise as in older children (Shi et al., 2011), potentially due to reduced image contrast. Some have opted to use T1 structural images from one child with longitudinal data as a template (Gao et al., 2015a, 2015b). Beyond this, a 4D template comprising both T1-weighted and T2-weighted images can reduce the bias introduced by image contrast as described earlier; however, this requires that laboratories collect both sequences (with sufficient image quality) at all stages along the developmental trajectory they wish to investigate. This can be costly and time-consuming. There are several 4D templates currently available (Oishi et al., 2018). However, none extend from birth to early childhood in a longitudinal sample, contain both T1 and T2 sequences, and have corresponding tissue priors. A close offering used by others (Graham et al., 2015a, 2015b; Graham et al., 2016) was a template containing both T1- and T2-weighted images from infants ages 0–4.5 years (3D templates for 0–2, 2–5, 5–8, 8–11, 11–14, 14–17, 17–21, 21–27, 27–33, 33–44, 44–60 months; Almli et al., 2007; Fonov et al., 2009;http://nist.mni.mcgill.ca/?p=1005). However, this template was only partially longitudinal and did not have corresponding tissue priors, which are critical for accurate segmentation. More recently, Zhang and colleagues (2016) generated a longitudinal 4D template containing T1- and T2-weighted images and corresponding tissue priors. However, this template currently extends only to 12 months of age (Zhang et al., 2016; https://www.nitrc.org/projects/infant_atlas_4d/. To our knowledge, no multi-year 4D templates for diffusion images have been released for the early developmental period, though a framework for constructing one has been proposed (Hart et al., 2010).
Suboptimal acquisition practices (e.g., sequence parameters; please see Data acquisition section above) for one developmental stage can also introduce bias if resulting image quality (e.g., due to low contrast or motion artifacts) is lower for children of that stage. This is of particular concern when registering images. For instance, typically, preprocessing procedures for functional imaging involve (1) co-registering functional and structural volumes, (2) normalizing structural volumes to a standard template, and then (3) applying the transformation in step 2 to the coregistered functional image. However, poor T1 or T2 image quality could lead to misalignment of functional and structural volumes in native space and/or misalignment of structural images with standard templates. The resulting functional images might therefore exhibit more spatial variability in the developmental stage with poorer image quality, which can subsequently affect second-level, age-related comparisons. Conversely, some laboratories have opted to perform functional analyses in native space, which eliminates bias introduced by normalization (Ferradal et al., 2018), but also prevents brain regions of different participants from occupying the same stereotaxic space, encumbering inter-subject comparisons and interpretations.
A similar bias can emerge for functional and diffusion imaging modalities specifically. For instance, realignment of functional volumes (i.e., registering all volumes in a time series to the first or mean volume) to control for head movement during a functional run can be less accurate for one developmental stage if the constituent images are of lower quality. Less accurate registration can also be expected for normalization if functional images are registered directly to a template (and not first co-registered with individual structural images, which are of higher contrast than functional images and therefore often preferred for normalization). In addition, a noisier signal can effect less precise estimation of task-evoked activation and functional connectivity. Lower quality scans can also disrupt diffusion pipelines, which may involve registration to subject-specific T1 or T2 images and to standard space templates for accurate fiber tract segmentation (Yeatman et al., 2012). Independent of misalignments and prior to tract segmentation, lower quality diffusion scans can also bias the quality of typical diffusion processing steps such as tensor fitting and fiber tractography, and optimal sequence parameters (e.g., b-values) may be different for infants and older cohorts (Bastiani et al., 2019).
4.3. Defining regions-of-interest and parcellations
To examine effects in specific brain regions or voxels, an investigator might define a region-of-interest (ROI). Anatomical parcellations are useful as ROIs because they reflect subdivisions of the entire brain according to structural boundaries. However, the challenges described above for templates also propagate to corresponding anatomical parcellations (for the templates that have corresponding parcellations). Parcellations corresponding to templates for older children cannot accurately label the anatomically smaller, and possibly spatially shifted, cortical and subcortical structures in infants and toddlers. Using age-specific parcellations can reduce these inaccuracies. Shi and colleagues (2011) have warped the AAL atlas from adults to infants (Shi et al., 2011), but this process still introduces bias as brain structures can shift in relative size and stereotaxic location to one another over the time course of brain development (Oishi et al., 2018), requiring visual inspection to confirm sufficient overlap between template and individual structures. It will be important for future anatomical parcellation methodologies to reflect the anatomy of younger children, rather than simply warped anatomy of older children or adults. Li and colleagues (2015) generated a 4D cortical surface atlas with volumes for chronical ages 1, 3, 6, 9, 12, 18, 24, 36, 48, 60, and 72 months using 339 longitudinally scanned MRIs from 50 healthy infants (Li et al., 2015b; https://www.nitrc.org/projects/infantsurfatlas; numbers updated from original report). However, this template and corresponding parcellation map only include the cortical surface and do not contain subcortical brain areas nor corresponding tissue priors. For a list of available infant parcellations current to 2018, please see (Oishi et al., 2018). To the best of our knowledge, there are currently no longitudinal templates from birth to approximately five years of age (even with annual scanning intervals) with corresponding tissue priors and anatomical parcellations. Lastly, like segmentation, manual delineation of structures is a time-consuming and labor-intensive, but effective, alternative (Vanderauwera et al., 2018).
ROIs to examine specific brain regions or networks may be defined in other ways, too. For instance, unlike anatomical parcellations, functional parcellations reflect brain regions subdivided according to their function (or functional connectivity) rather than structure (Arslan et al., 2018). Meanwhile, Brodmann areas represent brain regions according to cytoarchitecture. Longitudinal birth to two-year (Shi et al., 2018) and neonate (Scheinost et al., 2016) parcellations have been generated using resting-state functional connectivity data; however, these parcellations did not include older children, limiting developmental comparisons to younger children. Additionally, ROIs need not conform to a whole-brain subdivision, but can also be commonly constructed as representative spheres or cubes of voxels sampled from widespread parts of the brain. For instance, Smyser and colleagues (2016) warped infant resting-state data to an adult template and overlaid Power’s 264 ROIs, defined by homogeneity of BOLD signal fluctuations (Power et al., 2011). However, fifty ROIs did not overlap with infant gray matter (Smyser et al., 2016), again demonstrating that architecture varies considerably in stereotaxic location by developmental stages (even when mostly accounting for head size) and posing challenges for matching brain areas and networks from multiple stages. Overall, further research and tool development are needed to ensure unbiased parcellation and ROI selection across developmental stages.
Finally, coordinates are commonly used to characterize the location of brain effects. For children over five years of age and adults, individual brain images are often registered to templates in either Montreal Neurological Institute (MNI) and Talairach stereotaxic space, each of which come with standard coordinate systems. Any resulting brain effects may then be reported using MNI or Talairach coordinates; subsequent investigators can use these coordinates for ROI analyses after registering their own datasets to MNI or Talairach space. The lack of an accepted standardized coordinate system for infants is problematic (Akiyama et al., 2013), because if an investigator were to select ROIs for an infant study based on results of a previous infant study, then he or she would need to register the imaging data to the same template used by the previous study. Likewise, ROIs can be taken only from meta-analyses in older children and adults, but not infants, as quantitative meta-analytic techniques such as Activation Likelihood Estimation (Turkeltaub et al., 2002) are not possible without a standard coordinate system (or known conversions to such; Oishi et al., 2018).
4.4. Smoothing
Spatial smoothing, also considered blurring, is the process of averaging (weighted) intensities from nearby voxels. Smoothing increases SNR, but simultaneously decreases spatial specificity. The extent of the smoothing depends on the size of the smoothing kernel such that larger kernels increase SNR and decrease spatial specificity more than smaller kernels. As infant brains are considerably smaller than the brains of older children and adults (Groeschel et al., 2010), using kernels commonly used for older populations, such as 8 mm full width at half maximum (e.g., Yu et al., 2018), the default in SPM, could be problematic for localizing structures that are especially small in infants, such as the amygdala (Graham et al., 2015b). A recent study in infants younger than 10 days observed that the optimal compromise between SNR and spatial specificity occurred when using a smoothing kernel size of 3 mm full width at half maximum (Baxter et al., 2019); many infant studies have used a smoothing kernel size of 4 mm (Ferradal et al., 2018; Howell et al., 2019), 5 mm (Gabard-Durnam et al., 2018), or 6 mm (Fransson et al., 2007; Pruett et al., 2015; Merhar et al., 2016; Smyser et al., 2016; Mitra et al., 2017; Turesky et al., 2019). For developmental studies, use of smaller kernels for earlier developmental stages could standardize spatial specificity relative to brain size across stages. For instance, Gao and colleagues (2015) used increasing kernel sizes for increasing ages and head sizes, ranging from 4 mm for 1-month-old to 8.2 mm for 12-month-old infants (Gao et al., 2015a). While this adaptive approach may increase SNR in later compared with earlier developmental stages, it can also reduce differences in relative spatial specificity across developmental stages (Graham et al., 2015b). Accordingly, an adaptive approach to spatial smoothing may represent the best compromise if one selects kernel sizes that are considered to optimally balance SNR and spatial specificity for each developmental stage.
4.5. Head motion QC
Despite attempts during data acquisition to minimize participant motion, functional runs inevitably comprise head motion and the magnitude of this head motion can vary considerably, especially across developmental stages. Furthermore, for a given estimate of motion, relative to head size, this estimate would be greater for infants compared with later developmental stages. Head motion has been of major concern in developmental literature in children over five years of age, such that findings previously attributed to developmental changes (Fair et al., 2007, 2009; Dosenbach et al., 2010) were mitigated upon reanalysis with stricter motion criteria (Satterthwaite et al., 2012). The criteria for head motion have become extremely stringent in resting-state functional connectivity studies in infants, by comparison with early reports (Doria et al., 2010; Fransson et al., 2013). For instance, many studies now remove from functional time series data points in which the brain has moved from one volume to the next (i.e., framewise displacement) > 0.5 mm (Gao et al., 2015a, 2015b), > 0.3 mm (Graham et al., 2015a, 2015b; Graham et al., 2016; Rudolph et al., 2018), > 0.25 mm (Smyser et al., 2016) or > 0.2 mm (Pruett et al., 2015), sometimes along with additional data points that are temporally adjacent to the motion spike. A full run is often removed when the number of these artifactual volumes exceeds a particular threshold (e.g., 22.5 %;Graham et al., 2015a) or does not provide sufficient remaining data (e.g., five minutes of continuous data; Smyser et al., 2016). Importanty, the thresholds used for examination of functional connectivity are generally much more strict than for task-evoked activation. Lastly, some studies performing group comparisons have additionally ensured that the numbers of artifactual volumes removed do not differ between groups; otherwise, differences due to head motion could be mistakenly attributed to neuronal effects (Van Dijk et al., 2012). Although developmental neuroimaging practices in the 0−5 year age range might potentially benefit from this practice, further research is required to investigate the delicate trade-off between artifact removal and rejection of data that is extremely challenging to acquire in this population.
Additionally, in the calculation of framewise displacement (i.e., combined translation and rotation movement of the brain between contiguous volumes), an estimate of brain radius is often needed to convert rotational measures of degrees or radians to millimeters. Power and colleagues (2012) estimate 50 mm (Power et al., 2012); but this is presumably for the brain of an older child or adult, rather than an infant, which is considerably smaller (please see above and Table 1 for size comparisons). Apart from the question of consistent motion thresholds across ages, the field needs to establish whether an average head size estimate or different estimates for different ages should be used for the calculation of framewise displacement. Otherwise, framewise displacement could be assessed separately for translation and rotation, as is also fairly common in the literature (e.g., Yu et al., 2018), thereby obviating the need for conversion to millimeters using an estimate of head size. Though avoiding the concerns over head radius, this scenario would still be subject to the challenges described at the beginning of this subsection.
Head motion also hampers diffusion pipelines. Age-independent tools such as the FMRIB Software Library’s eddy tool (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/eddy) and DTIPrep (Oguz et al., 2014; https://www.nitrc.org/projects/dtiprepare) are often used in infants (Geng et al., 2012; Langer et al., 2017; Bastiani et al., 2019). Recently, a new motion correction tool called Spherical Harmonics and Radial Decomposition (SHARD; Christiaens et al., 2020; pre-print) was introduced for multi-shell high angular resolution diffusion imaging (HARDI) data, which carries more fine-grained microstructural information compared with classical diffusion imaging. Using a data-driven representation of multi-shell HARDI data, SHARD avoids imposing a model with numerous assumptions, and germane to the present review, has demonstrated efficacy for the challenging motion correction in infant datasets. However, this tool can be applied only when HARDI data are available. Ultimately, to control for motion across the 0−5 year age range, the field of developmental cognitive neuroscience will need to determine whether head motion thresholds should be the same or differentially strict across developmental stages.
4.6. fMRI specification
As described in the Data acquisition section above (please see Conditions for fMRI subsection), fMRI captures brain responses over time, most frequently through the BOLD signal. The BOLD signal, like other types of signals, can be decomposed (via a Fourier transform) and characterized by the frequencies that constitute it, with some frequencies constituting more of the signal than others. The degree to which a particular frequency constitutes a signal can be computed with power spectra analyses. Biases may also be introduced in the context of BOLD signal frequency. Power spectra analyses have revealed that the BOLD signal is dominated by different frequencies at different ages, with greatest representation for neonates at 0.0056 Hz compared with 0.028 Hz for one- and two-year-olds (Alcauter et al., 2015), likely with some variation depending on the number of points specified in the discrete Fourier transform. This has implications for resting-state, functional connectivity studies in infants, which often use band-pass filters with a lower threshold of roughly 0.008/9 Hz to remove artifacts or non-neurophysiological signal thought to manifest at these lower frequencies in adults. As such, these studies effectively remove the dominant signal for the neonate datasets (Gao et al., 2015a, 2015b;Graham et al., 2015a, 2015b; Pruett et al., 2015; Rudolph et al., 2018; Turesky et al., 2019). In a rare deviation, Graham et al. (2016) used a low-pass filter (< 0.1 Hz), which presumably would have retained these low frequencies (Graham et al., 2016), but perhaps low frequency artifacts, too. Future work needs to determine whether low frequency artifacts are present in infants as they are in older children, and if they are, whether their removal can be performed without removing potentially useful information from the lower frequencies of the BOLD signal. Lastly, heart rate and respiration rate are higher at younger ages, which should be considered when deciding on low-pass filters for removal of non-neurophysiological noise functional connectivity studies (Mongerson et al., 2017).
Developmental changes in vasculature (please see above) may also introduce age-related differences in neurovascular coupling (Arichi et al., 2012; Hagmann et al., 2012; Cusack et al., 2015; Grayson and Fair, 2017; Baxter et al., 2019), as has been investigated for children over five years of age and adults (Thomason et al., 2005). For instance, the hemodynamic response function (HRF), which characterizes changes in BOLD signal over time, was attenuated and exhibited a longer latency-to-peak in infants compared with adults for passive somatosensory responses (Arichi et al., 2012). Convolution of an age-appropriate HRF into the general linear model revealed activation in the somatosensory cortex that was not identified when convolving with the canonical adult HRF. Indeed, that HRF morphology and fit to stimulus-evoked responses affects the sensitivity of first-level modeling in infants (Baxter et al., 2019) and could generate false-positive effects in developmental comparisons of young children versus infants in task-evoked activation analyses. In other words, a poorly fit HRF would miss activation in the infants; then, when comparing to older children, it would appear as though only the older children exhibited activation in response to the stimulus, whereas the infants did not. Also, deactivation in infant primary sensory cortices has been observed in passive responses to auditory (Anderson et al., 2001) and visual stimuli (Born et al., 1998; Martin et al., 1999), suggesting that neurovascular coupling may be quantitatively and qualitatively different in infants compared with older ages. In addition, scanner background noise may have a bearing on infants’ auditory (and potentially visual) responses, as the neural processes underlying inhibition of scanner background noise may differ between infants and older children. Several approaches have been suggested to mitigate differences in hemodynamic responses between older children (age range: 7–12 years) and adults (Thomason et al., 2005), which broadly focus on ways to account for increased signal noise in children, and it is possible that some of these approaches can be applied to the early developmental age range as well.
In resting-state studies, the relationship between spontaneous fluctuations in hemodynamic signal and neural activity has also been investigated in adult mice (Mateo et al., 2017; Winder et al., 2017); but to our knowledge, this relationship remains to be characterized in young pups. Therefore, it is not entirely clear whether estimates of functional connectivity can similarly (as with estimates of fit in task-evoked designs) be affected by developmental differences in neurovascular coupling. Overall, neurovascular coupling across the 0−5 age range needs further elucidation (Hagmann et al., 2012) and quantification.
4.7. Controlling for age variance
When performing categorical comparisons between or among developmental stages, within-group variance in age also merits consideration. Specifically, the rapid neuroanatomical and neurophysiological changes early in development (Gilmore et al., 2018) can cause comparable age-related variances at early and late developmental stages to have markedly different consequences; e.g., ± two months when the child is four months represents greater relative variance compared with ± two months when the child is four years. Adjusting variables of interest (e.g., tissue volumes, functional connectivity estimates) for within-group variance in age, therefore, may improve second-level models.
5. Decisions for the field
Rapid changes to brain anatomy and physiology between birth and age five pose dozens of challenges for developmental MRI studies (Fig. 2). Acquisition and processing for children of different ages are often optimized using different tools, techniques, and parameters, which can lead to bias when conducting developmental comparisons. Broadly, this review has underscored and described the acquisition, processing, and analysis steps where these biases may emerge. For each step, the field must decide whether to optimize methods for the age-specific neurobiology, preserve methods across developmental stages, or employ an intermediate of these options.
Specific to data acquisition, we encourage the field to recognize that different states of consciousness can bias data interpretation. We urge the field to recommend whether voxel sizes should scale up with age and, for structural scans, whether infants should be scanned with T2 parameters while older children undergo T1 imaging, or whether we should use identical sequence parameters for all children. We urge the field to recommend how to account for differences in head coil fit and scanner and equipment upgrades during longitudinal studies, especailly since upgrades can improve image quality. For fMRI, we must address age-appropriatness of scanner tasks, including switches from passive to active tasks and difficulty levels. For determinations of sample size calculations, we need to take into account that higher attrition and data loss are likely for children 0−5 years of age, compared with developmental stages beyond five years.
Specific to data processing and analysis, the field needs to recommend whether to perform segmentation and image registration using age-appropriate tools, templates, tissue priors, and intensity inhomogeneity correction, or using age-independent practices. Notwithstanding, we encourage the field to acknowledge that tissue misclassifications and image misalignments will occur more frequently in earlier developmental stages and developmental stages scanned with age-inappropriate acquisition practices. The field also needs to recommend whether to use age-specific ROIs and parcellations, which capture age-specific structural and functional architecture, or a standard set of ROIs and parcellations that are subsequently registered to each developmental stage. Additionally, the field needs to recommend whether to smooth brain images with kernels that optimize SNR, spatial specificity, or the compromise between SNR and spatial specificity for each age group, or to smooth with identically sized kernels regardless of the developmental stage. We also must decide whether to control for head motion using thresholds that are commensurate with the degree of head motion, head size, and age of the child, or use identical thresholds across developmental stages. For fMRI, the field needs to recommend whether to employ age-appropriate frequency filters and HRFs. For statistical models, we should adjust variables of interest for within-group variance in age when performing comparisons across developmental stages.
Importantly, considerable care needs to be taken if employing age-specific approaches, to ensure that they are optimized for the anatomy and physiology unique to the age. We suggest two main approaches. The first is to rely on independently tested and reported methods that have been proven to be reliable for a particular age group. This approach could be useful for new investigators, who could follow the methodological approaches carefully developed, tested, and evaluated by another lab. If no “best practices” exist for the age of interest, then investigators need to adjust the parameters of their pipelines and carefully evaluate the fit and quality of the applied methods subject-by-subject. There are already a number of tools that can aid in determining success; e.g., Qoala-T rates tissue labelling from FreeSurfer segmentations (Klapwijk et al., 2019), though investigators may need to train its algorithms using a local dataset. In the absence of tools, independent raters can serve as an effective alternative.
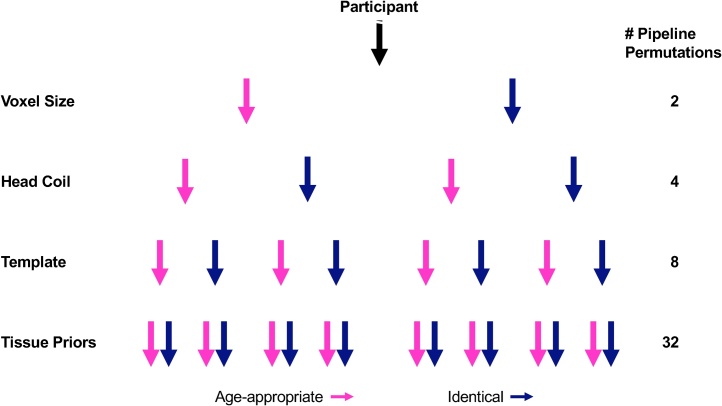
Overall, the field must decide on best practices to mitigate bias in developmental comparisons within individual studies. Standardizing these practices will also improve reproducibility across studies. Complex pipelines, such as those implemented in developmental studies, present investigators with acquisition, processing, analysis, and interpretation options at multiple stages, exponentially increasing the number of pipeline permutations (Fig. 3) and hindering reproducibility (Poldrack et al., 2017a, 2017b; Karayumak et al., 2019), which has become of major concern as part of the Open Science and Reproducibility movement (Poldrack et al., 2017a, 2017b; https://reproducibility.stanford.edu/about-us/). Importantly, these permutations may differ by study; therefore, the field must also clarify the contexts that would justify alternative pipelines (e.g., investigators may use smaller voxel sizes for younger compared with older children for structural, but not functional, pipelines). Ultimately, dissemination of guidelines should prune pipeline permutations and hopefully increase reproducibility.
Fig. 3.
Abbreviated hypothetical decision tree. For multiple stages of a developmental study, investigators must choose between age-appropriate methods (pink) or methods that are identical across developmental stages (blue), exponentially increasing the number of pipeline permutations. Please note that we have limited pipeline permutations here to four stages and binary options (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
One final consideration is that the large-scale, known neuroanatomical differences between early developmental stages (e.g., Matsuzawa et al., 2001; Knickmeyer et al., 2008; Groeschel et al., 2010; Sanchez et al., 2011; Geng et al., 2012; Gilmore et al., 2012; Li et al., 2013; Holland et al. 2014) underlie the utility of different methods to optimize images acquired in different developmental stages, and ultimately drive the trade-off between age-optimized methods and standardized methods across developmental stages. However, the experiments that demonstrate these large-scale neurodevelopmental changes were inevitably conducted in a manner that did not—and perhaps could not—completely eliminate bias, and it is unclear to what extent these biases may have affected reported results. Accordingly, circular reasoning emerges as our understanding of how to measure the brain accurately and unbiasedly has been based in part on results that were acquired with methods that also suffered from these biases.
6. Conclusion
The first years of life are marked by the most rapid and widespread changes in brain anatomy and physiology (Gilmore et al., 2018), with genetic or environmental experiences during this time shaping or altering typical developmental trajectories (Nelson and Gabard-durnam, 2020). Several laboratories have begun to use MRI to elucidate these trajectories (e.g., Matsuzawa et al., 2001; Knickmeyer et al., 2008; Geng et al., 2012; Gilmore et al., 2012; Li et al., 2013; Holland et al., 2014), but the rapid growth during this period poses substantial challenges for future developmental studies with methodological bias emerging in several places throughout data acquisition, processing, and analysis. One consistent theme of these specific challenges relates to a trade-off between optimizing approaches for each developmental stage, which inherently requires that multiple methods be used, or maintaining the same approaches across stages, which would likely alter the accuracy and precision in the estimates of brain structure or function across developmental stages. While the present review focused on the evaluation of different challenges for the field of developmental neuroimaging, further research is required for the construction of guidelines and approaches that are well-suited for the population of children ages 0−5 years. In addition, the utility of MRI in elucidating developmental trajectories is limited; consequently, there is a need for analogous descriptions of challenges for other modalities such as EEG, MEG, and fNIRS. We also encourage others to consider similar “calls to action” and to develop recommendations for the inchoate field of prenatal neuroimaging (Van Den and Thomason, 2016) and for infants born preterm, whose brain structure has been shown to differ from infants born at term (Giménez et al., 2008). To conclude, this review aims to underscore the challenges of developmental MRI studies of early childhood and to inform subsequent resolutions to these challenges.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank our funding sources for enabling us to write this review: Eunice Kennedy Shriver National Institute of Child Health and Human DevelopmentR01HD65762, Jacobs Advanced Research Fellowship, and NIH/Harvard Medical School Catalyst5UL1RR025758 (all awarded to NG). We also thank Ola Ozernov-Palchik, Xi Yu, Jennifer Zuk, Jade Dunstan, Carolyn King, Kathryn Garrisi, and Joseph Sanfilippo for conceptual feedback and additional edits.
References
- Akiyama L.F., Richards T.R., Imada T., Dager S.R., Wroblewski L., Kuhl P.K. vol. 8. 2013. pp. 1–9. (Age-Specific Average Head Template for Typically Developing 6-Month-Old Infants). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcauter S., Lin W., Smith J.K., Goldman B.D., Reznick J.S., Gilmore J.H., Gao W. Frequency of spontaneous BOLD signal shifts during infancy and correlates with cognitive performance. Dev. Cogn. Neurosci. 2015;12:40–50. doi: 10.1016/j.dcn.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli C.R., Rivkin M.J., Mckinstry R.C. The NIH MRI study of normal brain development (Objective-2): newborns, infants, toddlers, and preschoolers. Hum Brain Mapp J. 2007;35:308–325. doi: 10.1016/j.neuroimage.2006.08.058. [DOI] [PubMed] [Google Scholar]
- Altaye M., Holland S.K., Wilke M., Gaser C. Infant brain probability templates for MRI segmentation and normalization. Neuroimage. 2008;43:721–730. doi: 10.1016/j.neuroimage.2008.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A.W., Marois R., Colson E.R., Peterson B.S., Duncan C.C., Ehrenkranz R.A. Vol. 19. 2001. pp. 1–5. (Neonatal Auditory Activation Detected by Functional Magnetic Resonance Imaging). [DOI] [PubMed] [Google Scholar]
- Antonov N.K., Ruzal-shapiro C.B., Morel K.D., Millar W.S., Kashyap S., Lauren C.T., Garzon M.C. 2017. Feed and Wrap MRI Technique in Infants. [DOI] [PubMed] [Google Scholar]
- Arichi T., Fagiolo G., Varela M., Melendez-Calderon A., Allievi A., Merchant N., Tusor N., Counsell S.J., Burdet E., Beckmann C.F., Edwards A.D. Development of BOLD signal hemodynamic responses in the human brain. Neuroimage. 2012;63:663–673. doi: 10.1016/j.neuroimage.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan S., Makropoulos A., Robinson E.C., Rueckert D., Parisot S. NeuroImage Human brain mapping : a systematic comparison of parcellation methods for the human cerebral cortex. Neuroimage. 2018;170:5–30. doi: 10.1016/j.neuroimage.2017.04.014. [DOI] [PubMed] [Google Scholar]
- Ashburner J., Friston K.J. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bastiani M., Andersson J.L.R., Cordero-grande L., Murgasova M., Hutter J., Price A.N., Makropoulos A., Fitzgibbon S.P., Hughes E., Rueckert D., Victor S., Rutherford M., Edwards A.D., Smith S.M., Tournier J., Hajnal J.V., Jbabdi S., Sotiropoulos S.N. NeuroImage Automated processing pipeline for neonatal diffusion MRI in the developing Human Connectome Project. Neuroimage. 2019;185:750–763. doi: 10.1016/j.neuroimage.2018.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter L., Fitzgibbon S., Moultrie F., Goksan S., Jenkinson M., Smith S., Andersson J., Duff E., Slater R. NeuroImage Optimising neonatal fMRI data analysis : design and validation of an extended dHCP preprocessing pipeline to characterise noxious-evoked brain activity in infants. Neuroimage. 2019;186:286–300. doi: 10.1016/j.neuroimage.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare R.J., Chen J., Kelly C.E., Alexopoulos D., Smyser C.D., Rogers C.E., Loh W.Y., Matthews L.G., Cheong J.L.Y., Spittle A.J., Anderson P.J., Doyle L.W., Inder T.E., Seal M.L., Thompson D.K. Neonatal brain tissue classification with morphological adaptation and unified segmentation. Front. Neuroinform. 2016;10:1–17. doi: 10.3389/fninf.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born P., Leth H., Miranda M.J., Rostrup E., Stensgaard A., Peitersen B., Larsson H.B.W., Lou H.C. Visual activation in infants and young children studied by functional magnetic resonance imaging. Pediatr. Res. 1998;44:578–583. doi: 10.1203/00006450-199810000-00018. [DOI] [PubMed] [Google Scholar]
- Born A., Miranda M., Rostrup E., Toft P., Peitersen B., Larsson H., Lou H. Functional magnetic resonance imaging of the normal and abnormal visual system in early life. Neuropediatrics. 2000;31:24–32. doi: 10.1055/s-2000-15402. [DOI] [PubMed] [Google Scholar]
- Buckner R.L., Hrienen F.M., Yeo T.B.T. Opportunities and limitations of intrinsic functional connectivity MRI. Nat. Rev. Neurosci. 2013;16:832–837. doi: 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2001. Data Table of Infant Head Circumference-for-Age Charts. [Google Scholar]
- Christiaens D., Cordero-grande L., Pietsch M., Hutter J., Price A.N., Hughes E.J., Vecchiato K., Deprez M., Edwards A.D., Hajnal J.V., Tournier J. 2020. Scattered Slice SHARD Reconstruction for Motion Correction in Multi-shell Diffusion MRI. arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho-Santos V., Shih A.Y. Postnatal development of cerebrovascular structure and the neurogliovascular unit. WIREs Dev Biol. 2019;e363:1–20. doi: 10.1002/wdev.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M.W., Bassett D.S., Power J.D., Braver T.S., Petersen S.E. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83:238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counsell S.J., Rutherford M.A. 2002. Magnetic Resonance Imaging of the Newborn Brain. [Google Scholar]
- Courchesne E., Chisum H.J., Townsend J., Cowles A., Covington J., Egaas B., Harwood M., Hinds S., Press G.A., Address J.T. Normal brain development and aging: quantitative analysis at in vivo MR. Neuroradiology. 2000;216:672–682. doi: 10.1148/radiology.216.3.r00au37672. [DOI] [PubMed] [Google Scholar]
- Cusack R., Wild C., Linke A.C., Arichi T., Lee D.S.C., Han V.K. Optimizing stimulation and analysis protocols for neonatal fMRI. PLoS One. 2015;10:1–13. doi: 10.1371/journal.pone.0120202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Shi F., Wang L., Wu G. iBEAT : a toolbox for infant brain magnetic resonance image processing. Neuroinformatics. 2013;11:211–225. doi: 10.1007/s12021-012-9164-z. [DOI] [PubMed] [Google Scholar]
- de Macedo Rodrigues K., Ben-Avi E., Sliva D.D., Choe M., Drottar M., Wang R., Fischl B., Grant P.E., Zöllei L. A FreeSurfer-compliant consistent manual segmentation of infant brains spanning the 0–2 year age range. Front. Hum. Neurosci. 2015;9:1–12. doi: 10.3389/fnhum.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D.C., Dirks H., Muircheartaigh J.O., Walker L., Jerskey B.A., Lehman K., Han M., Waskiewicz N., Deoni S.C.L. Pediatric neuroimaging using magnetic resonance imaging during non-sedated sleep. Pediatr. Radiol. 2014;44:64–72. doi: 10.1007/s00247-013-2752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deen B., Richardson H., Dilks D.D., Takahashi A., Keil B., Wald L.L., Kanwisher N., Saxe R. Organization of high-level visual cortex in human infants. Nat. Commun. 2017;8:1–10. doi: 10.1038/ncomms13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deoni S.C.L., Dean D.C., Muircheartaigh J.O., Dirks H., Jerskey B.A. NeuroImage Investigating white matter development in infancy and early childhood using myelin water faction and relaxation time mapping. Neuroimage. 2012;63:1038–1053. doi: 10.1016/j.neuroimage.2012.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi C.N., Chandrasekharan A., Sundararaman V.K., Alex Z.C. Neonatal brain MRI segmentation : a review. Comput. Biol. Med. 2015;64:163–178. doi: 10.1016/j.compbiomed.2015.06.016. [DOI] [PubMed] [Google Scholar]
- Dhankhar S., Tyagi S., Prasad T. Brain MRI segmentation using K-means algorithm. National Conference on Advances in Knowledge Management. 2010:1–5. [Google Scholar]
- Doria V., Beckmann C.F., Arichi T., Merchant N., Groppo M., Turkheimer F.E., Counsell S.J., Murgasova M., Aljabar P., Nunes R.G., Larkman D.J., Rees G., Edwards A.D. Emergence of resting state networks in the preterm human brain. Proc. Natl. Acad. Sci. 2010;107:20015–20020. doi: 10.1073/pnas.1007921107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U.F., Nardos B., Cohen A.L., Fair D.A., Power J.D., Church J.A., Nelson S.M., Wig G.S., Vogel A.C., Lessov-schlaggar C.N., Barnes K.A., Dubis J.W., Feczko E., Rebecca S., Jr J.R.P., Barch D.M., Petersen S.E., Bradley L. Prediction of individual brain maturity using fMRI published by : american Association for the advancement of science linked references are available on JSTOR for this article : prediction of individual brain maturity using fMRI. Science. 2010;329(80):1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J., Alison M., Counsell S.J., Benders M.J.N.L. MRI of the neonatal brain : a review of methodological challenges and neuroscienti fi c advances. J. Magn. Reson. Imaging. 2020:1–26. doi: 10.1002/jmri.27192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein W., Glover G., Hardy C., Redington R. The intrinsic signal-to-Noise ratio in NMR imaging. Magn. Reson. Med. 1986;3:604–618. doi: 10.1002/mrm.1910030413. [DOI] [PubMed] [Google Scholar]
- Ellis C., Skalaban L., Yates T., Bejjanki V., Cordova N., Turk-Browne N. Re-imagining fMRI for awake behaving infants. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-18286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair Da, Dosenbach N.U.F., Church Ja, Cohen A.L., Brahmbhatt S., Miezin F.M., Barch D.M., Raichle M.E., Petersen S.E., Schlaggar B.L. Development of distinct control networks through segregation and integration. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13507–13512. doi: 10.1073/pnas.0705843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair D.A., Cohen A.L., Power J.D., Dosenbach N.U.F., Church J.A., Miezin F.M., Schlaggar B.L., Petersen S.E. Functional brain networks develop from a ‘“Local to distributed”’ organization. PLoS Comput. Biol. 2009;5:1–16. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferradal S.L., Gagoski B., Jaimes C., Yi F., Carruthers C., Vu C., Litt J.S., Larsen R., Sutton B., Grant P.E., Zöllei L. System-specific patterns of thalamocortical connectivity in early brain development as revealed by structural and functional MRI. Cereb. Cortex. 2018;29:1218–1229. doi: 10.1093/cercor/bhy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonov V., Evans A., McKinstry R., Almli C., Collins D. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage. 2009;47:S102. [Google Scholar]
- Fransson P., Skiold B., Horsch S., Nordell A., Blennow M., Lagercrantz H., Aden U. Resting-state networks in the infant brain. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15531–15536. doi: 10.1073/pnas.0704380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P., Skiold B., Engstro M., Hallberg B., Mosskin M., Åden U., Fransson P., Skio B., Lagercrantz H., Blennow M. Spontaneous brain activity in the newborn brain during natural sleep — an fMRI study in infants born at full term. Pediatr. Res. 2009;66:301–305. doi: 10.1203/PDR.0b013e3181b1bd84. [DOI] [PubMed] [Google Scholar]
- Fransson P., Åden U., Blennow M., Lagercrantz H. The functional architecture of the infant brain as revealed by resting-state fMRI. Cereb. Cortex. 2011;21:145–154. doi: 10.1093/cercor/bhq071. [DOI] [PubMed] [Google Scholar]
- Fransson P., Metsaranta M., Blennow M., Ulrika A., Lagercrantz H., Vanhatalo S. Early Development of Spatial Patterns of Power-Law Frequency Scaling in fMRI Resting- State and EEG Data in the Newborn Brain. Cereb. Cortex. 2013;23:638–646. doi: 10.1093/cercor/bhs047. [DOI] [PubMed] [Google Scholar]
- Gaab N., Gaser C., Zaehle T., Jancke L., Schlaug G. Functional anatomy of pitch memory — an fMRI study with sparse temporal sampling. Neuroimage. 2003;19:1417–1426. doi: 10.1016/s1053-8119(03)00224-6. [DOI] [PubMed] [Google Scholar]
- Gaab N., Gabrieli J.D.E., Glover G.H. Assessing the Influence of Scanner Background Noise on Auditory Processing. I. An fMRI Study Comparing Three Experimental Designs with Varying Degrees of Scanner Noise. Hum. Brain Mapp. 2007;28:703–720. doi: 10.1002/hbm.20298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaab N., Gabrieli J.D.E., Glover G.H. Assessing the Influence of Scanner Background Noise on Auditory Processing. II. An fMRI Study Comparing Auditory Processing in the Absence and Presence of Recorded Scanner Noise Using a Sparse Design. Hum. Brain Mapp. 2007;28:721–732. doi: 10.1002/hbm.20299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaab N., Gabrieli J.D.E., Glover G.H. Resting in peace or noise : scanner background noise suppresses default-mode network. Hum. Brain Mapp. 2008;29:858–867. doi: 10.1002/hbm.20578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabard-Durnam L.J., Muircheartaigh J.O., Dirks H., Dean D., Tottenham N., Deoni S. Human amygdala functional network development : a cross-sectional study from 3 months to 5 years of age. Dev. Cogn. Neurosci. 2018;34:63–74. doi: 10.1016/j.dcn.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Zhu H., Giovanello K., Smith J.K., Shen D., Gilmore J.H., Lin W. Evidence on the emergence of the brain’ s default network from 2-week-old to 2-year-old healthy pediatric subjects. Proc. Natl. Acad. Sci. 2009;106:6790–6795. doi: 10.1073/pnas.0811221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Alcauter S., Elton A., Hernandez-Castillo C.R., Smith J.K., Ramirez J., Lin W. Functional network development during the first year: relative sequence and socioeconomic correlations. Cereb. Cortex. 2015;25:2919–2928. doi: 10.1093/cercor/bhu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Alcauter S., Smith J.K., Gilmore J.H., Lin W. Development of human brain cortical network architecture during infancy. Brain Struct. Funct. 2015;220:1173–1186. doi: 10.1007/s00429-014-0710-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X., Gouttard S., Sharma A., Gu H., Styner M., Lin W., Gerig G., Gilmore J.H. NeuroImage Quantitative tract-based white matter development from birth to age 2 years. Neuroimage. 2012;61:542–557. doi: 10.1016/j.neuroimage.2012.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore J.H., Shi F., Woolson S.L., Knickmeyer R.C., Short S.J., Lin W., Zhu H., Hamer R.M., Styner M., Shen D. Longitudinal development of cortical and subcortical gray matter from birth to 2 years. Cereb. Cortex. 2012;22:2478–2485. doi: 10.1093/cercor/bhr327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore J., Knickmeyer R.C., Gao W. Imaging structural and functional brain development in early childhood. Nat. Rev. Neurosci. 2018;19:123–137. doi: 10.1038/nrn.2018.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez M., Miranda M.J., Born A.P., Nagy Z., Rostrup E., Jernigan T.L. Authors accelerated cerebral white matter development in preterm infants : a voxel-based morphometry study with diffusion tensor MR imaging. Neuroimage. 2008;41:728–734. doi: 10.1016/j.neuroimage.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Glover G.H., Mueller B.A., Turner J.A., Van Erp T.G.M., Liu T.T., Greve D.N., Voyvodic J.T., Rasmussen J., Brown G.G., Keator D.B., Calhoun V.D., Lee H.J., Ford J.M., Mathalon D.H., Diaz M., Leary D.S.O., Gadde S., Preda A., Lim K.O., Wible C.G., Stern H.S., Belger A. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. J. Magn. Reson. Imaging. 2012;54:39–54. doi: 10.1002/jmri.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A.M., Pfeifer J.H., Fisher P.A., Carpenter S., Fair D.A. Early life stress is associated with default system integrity and emotionality during infancy. J Child Psychol Psychiatry Allied Discip. 2015;56:1212–1222. doi: 10.1111/jcpp.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A.M., Pfeifer J.H., Fisher P.A., Lin W., Gao W., Fair D.A. The potential of infant fMRI research and the study of early life stress as a promising exemplar. Dev. Cogn. Neurosci. 2015;12:12–39. doi: 10.1016/j.dcn.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A.M., Buss C., Rasmussen J.M., Rudolph M.D., Demeter D.V., Gilmore J.H., Styner M., Entringer S., Wadhwa P.D., Fair D.A. Implications of newborn amygdala connectivity for fear and cognitive development at 6-months-of-age. Dev. Cogn. Neurosci. 2016;18:12–25. doi: 10.1016/j.dcn.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C., Laumann T.O., Nielsen A.N., Schlaggar B.L., Dosenbach N.U.F., Petersen S.E., Gratton C., Laumann T.O., Nielsen A.N., Greene D.J., Gordon E.M. Functional Brain Networks Are Dominated by Stable Group and Individual Factors, Not Cognitive or Daily Article Functional Brain Networks Are Dominated by Stable Group and Individual Factors. Not Cognitive or Daily Variation. 2018:439–452. doi: 10.1016/j.neuron.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson D.S., Fair D.A. Development of large-scale functional networks from birth to adulthood: a guide to the neuroimaging literature. Neuroimage. 2017;160:15–31. doi: 10.1016/j.neuroimage.2017.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeschel S., Vollmer B., King M.D., Connelly A. International Journal of Developmental Neuroscience Developmental changes in cerebral grey and white matter volume from infancy to adulthood. Int. J. Dev. Neurosci. 2010;28:481–489. doi: 10.1016/j.ijdevneu.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Hagmann P., Grant P.E., Fair D.A., Knösche T.R., Planck M. MR connectomics : a conceptual framework for studying the developing brain. Front. Syst. Neurosci. 2012;6:1–17. doi: 10.3389/fnsys.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb R., Whiteus C., Freitas C., Grutzendler J. In vivo imaging of cerebral microvascular plasticity from birth to death. J. Cereb. Blood Flow Metab. 2013;106815:146–156. doi: 10.1038/jcbfm.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G., Shi Y., Zhu H., Sanchez M., Styner M., Niethammer M. 2010. DTI Longitudinal Atlas Construction As an Average of Growth Models. PrePring. [Google Scholar]
- Holland D., Chang L., Ernst T.M., Curran M., Buchthal S.D., Alicata D., Skranes J., Johansen H., Hernandez A., Yamakawa R., Kuperman J.M., Dale A.M. Structural growth trajectories and rates of change in the first 3 months of infant brain development. JAMA Neurol. 2014;71:1266–1274. doi: 10.1001/jamaneurol.2014.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz S.G., Braun A.R., Carr W.S., Picchioni D., Balkin T.J., Fukunaga M., Duyn J.H. Decoupling of the brain’s default mode network during deep sleep. Proc Natl Acad Sci. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.R., Styner M.A., Gao W., Yap P., Wang L., Baluyot K., Yacoub E., Chen G., Potts T., Salzwedel A., Li G., Gilmore J.H., Piven J., Smith J.K., Shen D., Ugurbil K., Zhu H., Lin W., Elison J.T. The UNC / UMN Baby Connectome Project (BCP): an overview of the study design and protocol development. Neuroimage. 2019;185:891–905. doi: 10.1016/j.neuroimage.2018.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E.J., Winchman T., Padormo F., Teixeira R., Wurie J., Sharma M., Fox M., Hutter J., Cordero-grande L., Price A.N., Allsop J., Bueno-conde J., Tusor N., Arichi T., Edwards A.D., Rutherford M.A., Counsell S.J., Hajnal J.V. A dedicated neonatal brain imaging system. Magn. Reson. Med. 2017;804:794–804. doi: 10.1002/mrm.26462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim T., Few K., Greenwood R., Smith C., Malcolm P., Johnson G., Lally P., Thayyil S., Clarke P. ‘Feed and wrap’ or sedate and immobilise for neonatal brain MRI? Arch. Dis. Child. Fetal Neonatal Ed. 2015;100:465–466. doi: 10.1136/archdischild-2015-308847. [DOI] [PubMed] [Google Scholar]
- Karayumak S.C., Bouix S., Ning L., James A., Crow T., Shenton M., Kubicki M., Rathi Y. NeuroImage Retrospective harmonization of multi-site diffusion MRI data acquired with different acquisition parameters. Neuroimage. 2019;184:180–200. doi: 10.1016/j.neuroimage.2018.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil B., Alagappan V., Mareyam A., Mcnab J.A., Fujimoto K., Tountcheva V., Triantafyllou C., Dilks D.D., Kanwisher N., Lin W., Grant P.E., Wald L.L. Size-optimized 32-Channel brain arrays for 3 t pediatric imaging. Magn. Reson. Med. 2011;66:1777–1787. doi: 10.1002/mrm.22961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapwijk E.T., Van De Kamp F., Van Der Meulen M., Peters S., Wierenga L.M. Qoala-T : a supervised-learning tool for quality control of FreeSurfer segmented MRI data. Neuroimage. 2019;189:116–129. doi: 10.1016/j.neuroimage.2019.01.014. [DOI] [PubMed] [Google Scholar]
- Knickmeyer R.C., Gouttard S., Kang C., Evans D., Wilber K., Smith J.K., Hamer R.M., Lin W., Gerig G., Gilmore J.H. A structural MRI study of human brain development from birth to 2 years. J. Neurosci. 2008;28:12176–12182. doi: 10.1523/JNEUROSCI.3479-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer N., Peysakhovich B., Zuk J., Drottar M., Sliva D.D., Smith S., Becker B.L.C., Grant P.E., Gaab N. White Matter Alterations in Infants at Risk for Developmental Dyslexia. Cereb. Cortex. 2017;27:1027–1036. doi: 10.1093/cercor/bhv281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Nie J., Wang L., Shi F., Lin W., Gilmore J.H., Shen D. Mapping region-specific longitudinal cortical surface expansion from birth to 2 years of age. Cereb. Cortex. 2013;23:2724–2733. doi: 10.1093/cercor/bhs265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wang L., Shi F., Gilmore J.H., Lin W., Shen D. Construction of 4D high-definition cortical surface atlases of infants : methods and applications. Med. Image Anal. 2015;25:22–36. doi: 10.1016/j.media.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Park B., Liu W., Zhang J., Reed M.P., Rupp J.D., Hoff C.N., Hu J. A statistical skull geometry model for children 0-3 years old. PLoS. 2015:1–13. doi: 10.1371/journal.pone.0127322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.C., Flax J.F., Guise K.G., Sukul V., Benasich A.A. Functional connectivity of the sensorimotor area in naturally sleeping infants. Brain Res. 2008;1223:42–49. doi: 10.1016/j.brainres.2008.05.054. [DOI] [PubMed] [Google Scholar]
- Lyall A.E., Shi F., Geng X., Woolson S., Li G., Wang L., Hamer R.M., Shen D., Gilmore J.H. 2015. Dynamic Development of Regional Cortical Thickness and Surface Area in Early Childhood; pp. 2204–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhyastha T., Peverill M., Koh N., Mccabe C., Flournoy J., Mills K., King K., Pfeifer J., Mclaughlin K.A. Developmental Cognitive Neuroscience Current methods and limitations for longitudinal fMRI analysis across development. Dev. Cogn. Neurosci. 2018;33:118–128. doi: 10.1016/j.dcn.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makropoulos A., Gousias I.S., Ledig C., Aljabar P., Serag A., Hajnal J.V., Edwards A.D., Counsell S.J., Rueckert D. Automatic whole brain MRI segmentation of the developing neonatal brain. IEEE Trans. Med. Imaging. 2014;33:1818–1831. doi: 10.1109/TMI.2014.2322280. [DOI] [PubMed] [Google Scholar]
- Makropoulos A., Counsell S.J., Rueckert D. NeuroImage A review on automatic fetal and neonatal brain MRI segmentation. Neuroimage. 2018;170:231–248. doi: 10.1016/j.neuroimage.2017.06.074. [DOI] [PubMed] [Google Scholar]
- Martin E., Joeri P., Loenneker T., Ekatodramis D., Vitacco D., Hennig J. Visual processing in infants and children studied using functional MRI. Pediatr. Res. 1999;46:135–140. doi: 10.1203/00006450-199908000-00001. [DOI] [PubMed] [Google Scholar]
- Mateo C., Knutsen P.M., Tsai P.S., Shih A.Y., Kleinfeld D. Entrainment of arteriole vasomotor fluctuations by neural activity is a basis of blood-oxygenation-Level-Dependent “Resting-State” connectivity. Neuron. 2017;96:936–948. doi: 10.1016/j.neuron.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa J., Matsui M., Konishi T. Age-related Volumetric Changes of Brain Gray and White Matter in Healthy Infants and Children. Cereb. Cortex. 2001;11:335–342. doi: 10.1093/cercor/11.4.335. [DOI] [PubMed] [Google Scholar]
- Merhar S.L., Gozdas E., Tkach J.A., Harpster K.L., Schwartz T.L., Yuan W., Kline-Fath B.M., Leach J.L., Altaye M., Holland S.K. Functional and structural connectivity of the visual system in infants with perinatal brain injury. Pediatr. Res. 2016;80:43–48. doi: 10.1038/pr.2016.49. [DOI] [PubMed] [Google Scholar]
- Mitra A., Snyder A.Z., Tagliazucchi E., Laufs H., Elison J., Emerson R.W., Shen M.D., Wolff J.J., Botteron K.N., Dager S., Estes A.M., Evans A., Gerig G., Hazlett H.C., Paterson S.J., Schultz R.T., Styner M.A., Zwaigenbaum L., Network T.I. 2017. Resting-state fMRI in Sleeping Infants More Closely Resembles Adult Sleep Than Adult Wakefulness; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongerson C.R.L., Jennings R.W., Borsook D., Becerra L., Bajic D. Resting-state functional connectivity in the infant brain: methods, pitfalls, and potentiality. Front. Pediatr. 2017;5:1–26. doi: 10.3389/fped.2017.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.A., Gabard-durnam L.J. Early adversity and critical periods: neurodevelopmental consequences of violating the expectable environment. Trends Neurosci. 2020;43:133–143. doi: 10.1016/j.tins.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman M., O’Kusky J. The growth and development of microvasculature in human cerebral cortex. J. Neuropathol. Exp. Neurol. 1986;45:222–232. [PubMed] [Google Scholar]
- Ogawa S., Lee T.-M. Magnetic resonance imaging of blood vessels at high fields: in vivo and in vitro measurements and image simulation. Magn. Reson. Med. 1990;16:9–18. doi: 10.1002/mrm.1910160103. [DOI] [PubMed] [Google Scholar]
- Oguz I., Farzinfar M., Matsui J., Budin F., Liu Z., Gerig G., Johnson H.J., Styner M. DTIPrep : quality control of diffusion-weighted images. Front. Neuroinform. 2014;8:1–11. doi: 10.3389/fninf.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Chang L., Huang H. Baby brain atlases. Neuroimage. 2018:1–16. doi: 10.1016/j.neuroimage.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T., Collins D.L., Evans A.C., Leonard G., Pike B., Zijdenbos A. Maturation of white matter in the human brain : a review of magnetic resonance studies. Brain Res. Bull. 2001;54:255–266. doi: 10.1016/s0361-9230(00)00434-2. [DOI] [PubMed] [Google Scholar]
- Perrachione T.K., Ghosh S.S. Optimized design and analysis of sparse-sampling fMRI experiments. Front. Neurosci. 2013;7:1–18. doi: 10.3389/fnins.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A., Mathalon D.H., Sullivan E.V., Rawles J.M., Zipursky R.B., Lim K.O. A quantitative magnetic resonance imaging study of changes in brain morphology from infancy to late adulthood. Arch. Neurol. 1994;51:874–887. doi: 10.1001/archneur.1994.00540210046012. [DOI] [PubMed] [Google Scholar]
- Picchioni D., Duyn J.H., Horovitz S.G. Sleep and the functional connectomez.star; Neuroimage. 2013;80:387–396. doi: 10.1016/j.neuroimage.2013.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack R.A., Baker C.I., Durnez J., Gorgolewski K.J., Matthews P.M., Munaf M.R., Nichols T.E., Poline J., Vul E., Yarkoni T. Scanning the horizon : towards. Nat. Rev. Neurosci. 2017;18:115–126. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack R.A., Baker C.I., Durnez J., Gorgolewski K.J., Matthews P.M., Munafò M.R., Nichols T.E., Poline J.B., Vul E., Yarkoni T. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci. 2017;18:115–126. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomponio R., Erus G., Habes M., Doshi J., Srinivasan D., Mamourian E., Bashyam V., Nasrallah I.M., Satterthwaite T.D., Fan Y., Launer L.J., Masters C.L., Maruff P., Zhuo C., Johnson S.C., Fripp J., Koutsouleris N., Wolf D.H., Gur R., Gur R., Morris J., Albert M.S., Grabe H.J., Resnick S.M., Bryan R.N., Wolk D.A., Shinohara R.T., Shou H., Davatzikos C. NeuroImage Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. Neuroimage. 2020;208:1–15. doi: 10.1016/j.neuroimage.2019.116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Cohen A.L., Nelson S.M., Wig G.S., Barnes K.A., Church J.A., Vogel A.C., Laumann T.O., Miezin F.M., Schlaggar B.L., Petersen S.E. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., Petersen S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prastawa M., Gilmore J.H., Lin W., Gerig G. Automatic segmentation of MR images of the developing newborn brain. Med. Image Anal. 2005;9:457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Pruett J.R., Kandala S., Hoertel S., Snyder A.Z., Elison J.T., Nishino T., Feczko E., Dosenbach N.U.F., Nardos B., Power J.D., Adeyemo B., Botteron K.N., McKinstry R.C., Evans A.C., Hazlett H.C., Dager S.R., Paterson S., Schultz R.T., Collins D.L., Fonov V.S., Styner M., Gerig G., Das S., Kostopoulos P., Constantino J.N., Estes A.M., Petersen S.E., Schlaggar B.L., Piven J. Accurate age classification of 6 and 12 month-old infants based on resting-state functional connectivity magnetic resonance imaging data. Dev. Cogn. Neurosci. 2015;12:123–133. doi: 10.1016/j.dcn.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle M.E. Behind the scenes of functional brain imaging: a historical and physiological perspective. Proc. Natl. Acad. Sci. 1998;95:765–772. doi: 10.1073/pnas.95.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raininko R., Autti T., Vanhanen S.L., Ylikoskp A., Erkinjuntti T., Santavuori P. The normal brain stem from infancy to old age: a morphometric MRI study. Neuroradiology. 1994;36:364–368. doi: 10.1007/BF00612119. [DOI] [PubMed] [Google Scholar]
- Raschle N.M., Chang M., Gaab N. Structural brain alterations associated with dyslexia predate reading onset. Neuroimage. 2011;57:742–749. doi: 10.1016/j.neuroimage.2010.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle N., Zuk J., Gaab N. Functional characteristics of developmental dyslexia in left-hemispheric posterior brain regions predate reading onset. Proc Natl Acad Sci. 2012;109:2156–2161. doi: 10.1073/pnas.1107721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle N., Zuk J., Ortiz-Mantilla S., Sliva D.D., Franceschi A., Grant P.E., Benasich A.A., Gaab N. Pediatric neuroimaging in early childhood and infancy: challenges and practical guidelines. Ann. N. Y. Acad. Sci. 2012;1252:43–50. doi: 10.1111/j.1749-6632.2012.06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle N., Stering P.L., Meissner S.N., Gaab N. Altered neuronal response during rapid auditory processing and its relation to phonological processing in prereading children at familial risk for dyslexia. Cereb. Cortex. 2014;24:2489–2501. doi: 10.1093/cercor/bht104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath T.W. Signal-to-noise ratio in MRI. Br. J. Radiol. 1998;71:704–707. doi: 10.1259/bjr.71.847.9771379. [DOI] [PubMed] [Google Scholar]
- Reynolds J.E., Grohs M.N., Dewey D., Lebel C. NeuroImage Global and regional white matter development in early childhood. Neuroimage. 2019;196:49–58. doi: 10.1016/j.neuroimage.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Rudolph M.D., Graham A.M., Feczko E., Miranda-Dominguez O., Rasmussen J.M., Nardos R., Entringer S., Wadhwa P.D., Buss C., Fair D.A. Maternal IL-6 during pregnancy can be estimated from newborn brain connectivity and predicts future working memory in offspring. Nat. Neurosci. 2018;21:765–772. doi: 10.1038/s41593-018-0128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge V.M., Nitz W.R., Trelles M., Goerner F.L. 3rd ed. Thieme Medical Publishers Inc; 2013. Physics of Clinical MR Taught Through Images. [Google Scholar]
- Sanchez C.E., Richards J.E., Almli C.R. 2011. Neurodevelopmental MRI Brain Templates for Children From 2 Weeks to 4 Years of Age. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite T.D., Wolf D.H., Loughead J., Ruparel K., Elliott M.A., Hakonarson H., Gur R.C., Gur R.C.E. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite T.D., Elliott M.A., Gerraty R.T., Ruparel K., Loughead J., Calkins M.E., Eickhoff S.B., Hakonarson H., Gur R.C., Gur R.C.E., Wolf D.H. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders D.E., Thompson C., Gunny R., Jones R., Cox T. Magnetic resonance imaging protocols for paediatric neuroradiology. Pediatr. Radiol. 2007;37:789–797. doi: 10.1007/s00247-007-0462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinost D., Hyun S., Xilin K., Cheryl S. Preterm birth alters neonatal, functional rich club organization. Brain Struct. Funct. 2016;221:3211–3222. doi: 10.1007/s00429-015-1096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Davatzikos C. HAMMER : hierarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Shi F., Yap P.T., Wu G., Jia H., Gilmore J.H., Lin W., Shen D. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Salzwedel A.P., Lin W., Gilmore J.H. 2018. Functional Brain Parcellations of the Infant Brain and the Associated Developmental Trends; pp. 1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyser C.D., Dosenbach N.U.F., Smyser T.A., Snyder A.Z., Rogers C.E., Inder T.E., Schlaggar B.L., Neil J.J. Prediction of brain maturity in infants using machine-learning algorithms. Neuroimage. 2016;136:1–9. doi: 10.1016/j.neuroimage.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange J.P., Jenkins L.M., Bessette K.L., Kling L.R., Bark J.S., Shepard R., Hamlat E.J., Deldonno S., Phan K.L., Passarotti A.M., Ajilore O., Langenecker S.A. Predictors of Attrition in Longitudinal Neuroimaging Research: Brain Connect. Brain Connect. 2018;8:527–536. doi: 10.1089/brain.2018.0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague S., Youssef G.J., Macdonald J.A., Sciberras E., Shatte A., Fuller-tyszkiewicz M., Greenwood C., Mcintosh J., Olsson C.A. Retention strategies in longitudinal cohort studies : a systematic review and meta- analysis. BMC Med. Res. Methodol. 2018;18:1–22. doi: 10.1186/s12874-018-0586-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieba C., Frayne A., Walton M., Mah A., Benischeck A., Dewey D., Lebel C. Factors associated with successful MRI scanning in Unsedated Young Children. Front. Pediatr. 2018;6:1–5. doi: 10.3389/fped.2018.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason M.E., Burrows B.E., Gabrieli J.D.E., Glover G.H. Breath holding reveals differences in fMRI BOLD signal in children and adults. Neuroimage. 2005;25:824–837. doi: 10.1016/j.neuroimage.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Turesky T., Jensen S., Yu X., Kumar S., Wang Y., Sliva D., Gagoski B., Sanfilippo J., Zöllei L., Boyd E., Haque R., Kakon S., Islam N., Petri W., Nelson C., N G The relationship between biological and psychosocial risk factors and resting-state functional connectivity in 2-month-old Bangladeshi infants: a feasibility and pilot study. Dev. Sci. 2019;22:e12841. doi: 10.1111/desc.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turesky T., Xie W., Kumar S., Sliva D.D., Gagoski B., Vaughn J., Lilla Z., Petri W.A., Nelson C.A., Gaab N. Relating anthropometric indicators to brain structure in 2-month-old Bangladeshi infants growing up in poverty: a pilot study. Neuroimage. 2020;210 doi: 10.1016/j.neuroimage.2020.116540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeltaub P.E., Eden G.F., Jones K.M., Zeffiro T.A. Meta-analysis of the functional neuroanatomy of single-word reading: method and validation. Neuroimage. 2002;16:765–780. doi: 10.1006/nimg.2002.1131. [DOI] [PubMed] [Google Scholar]
- Tustison N.J., Avants B.B., Cook P.A., Zheng Y., Egan A., Yushkevich P.A., Gee J.C. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Heuvel M.I., Thomason M.E. Functional connectivity of the human brain in utero. Trends Cogn. Sci. (Regul. Ed.) 2016;20:931–939. doi: 10.1016/j.tics.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk K.R.A., Hedden T., Venkataraman A., Evans K.C., Lazar S.W., Buckner R.L. Intrinsic functional connectivity As a tool for human connectomics : theory, properties, and optimization. J. Neurophysiol. 2010;02138:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk K.R.A., Sabuncu M.R., Buckner R.L. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera J., Wouters J., Vandermosten M. Developmental Cognitive Neuroscience Early dynamics of white matter de fi cits in children developing dyslexia. Dev. Cogn. Neurosci. 2017;27:69–77. doi: 10.1016/j.dcn.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera J., Altarelli I., Vandermosten M., De Vos A., Wouters J., Ghesquière P. Atypical structural asymmetry of the planum temporale is related to family history of dyslexia. Cereb. Cortex. 2018;28:63–72. doi: 10.1093/cercor/bhw348. [DOI] [PubMed] [Google Scholar]
- Vasung L., Turk E.A., Ferradal S.L., Sutin J., Stout J.N., Ahtam B., Lin P., Grant P.E. Exploring early human brain development with structural and physiological neuroimaging. Neuroimage. 2019;187:226–254. doi: 10.1016/j.neuroimage.2018.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar N., Mills K.L., Alexander-bloch A., Tamnes C.K., Whittle S. Developmental Cognitive Neuroscience Structural brain development : a review of methodological approaches and best practices. Dev. Cogn. Neurosci. 2018;33:129–148. doi: 10.1016/j.dcn.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M., Schmithorst V.J., Holland S.K. Normative pediatric brain data for spatial normalization and segmentation differs from standard adult data. Magn. Reson. Med. 2003;50:749–757. doi: 10.1002/mrm.10606. [DOI] [PubMed] [Google Scholar]
- Winder A.T., Echagarruga C., Zhang Q., Drew P.J. Weak correlations between hemodynamic signals and ongoing neural activity during the resting state. Nat. Neurosci. 2017;20:1761–1769. doi: 10.1038/s41593-017-0007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Avants B. Automatic registration-based segmentation for neonatal brains using ANTs and Atropos. MICCAI Gd Chall Neonatal Brain Segmentation. 2012;36 [Google Scholar]
- Yeatman J.D., Dougherty R.F., Myall N.J., Wandell B.A., Feldman H.M. Tract profiles of white matter properties: automating fiber-tract quantification. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Li T., Hung S., Zhang H., Wang L., Shen D., Zhu H. The emergence of a functionally flexible brain during early infancy. Proc. Natl. Acad. Sci. 2020;117:23904–23913. doi: 10.1073/pnas.2002645117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Schummers J., Sur M. The influence of astrocyte activation on hemodynamic signals for functional brain imaging. In: Roe A., editor. Imaging the Brain With Optical Methods. Springer Science+Business Media; 2010. [Google Scholar]
- Yu X., Raney T., Perdue M.V., Zuk J., Ozernov-palchik O., Becker B.L.C., Raschle N.M., Gaab N. Emergence of the neural network underlying phonological processing from the prereading to the emergent reading stage: a longitudinal study. Hum. Brain Mapp. 2018;39:2047–2063. doi: 10.1002/hbm.23985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr L., de Traversay J. Premature infant re- sponses to noise reduction by earmuffs: effects on behavioral and physiologic measures. J. Perinatol. 1995;15:448–455. [PubMed] [Google Scholar]
- Zhang Y., Shi F., Wu G., Wang L., Yap P., Shen D. Consistent spatial-temporal longitudinal atlas construction for developing infant brains. IEEE Trans. Med. Imaging. 2016;35:2568–2577. doi: 10.1109/TMI.2016.2587628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Shen D., Lin W. Resting-state functional MRI studies on infant brains: a decade of gap-filling efforts han. Neuroimage. 2019;185:664–684. doi: 10.1016/j.neuroimage.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllei L., Iglesias J.E., Ou Y., Grant P.E., Fischl B. Infant FreeSurfer: an automated segmentation and surface extraction pipeline for T1-weighted neuroimaging data of infants 0-2 years. Neuroimage. 2020 doi: 10.1016/j.neuroimage.2020.116946. [DOI] [PMC free article] [PubMed] [Google Scholar]