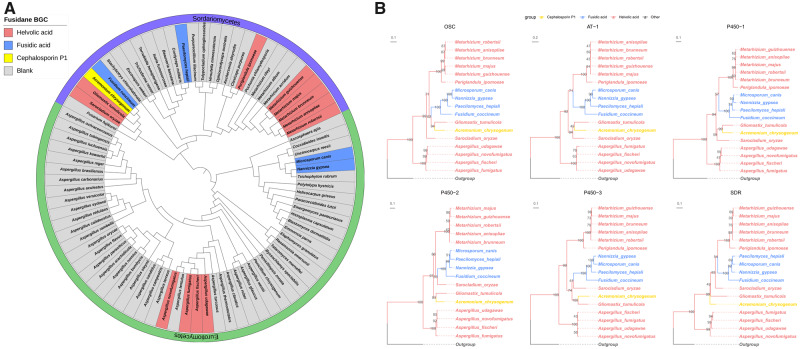
Fig. 2.
Maximum likelihood phylograms of fusidane BGCs and their host species. (A) The taxonomic status of the 17 fusidane species and their relatives. The rooted tree was determined from the protein sequence of 83 BUSCOs. Saccharomyces cerevisiae was set as outgroup and pruned subsequently. The scale bar indicates the mean expected substitutions per site. (B) Rooted phylogenetic trees of OSC, AT-1, P450-1, P450-2, P450-3, and SDR genes. Outgroups were used to root these trees: ORX70593.1 and ORX69994.1 from Linderina pennispora for OSC and AT-1; KEY83654.1 from Aspergillus fumigatus for P450-1; XP_751354.1 and XP_751350.1 from A. fumigatus for P450-2 and P450-3; and KZZ87816.1 from Moelleriella libera for SDR, respectively. The best evolutionary model for the construction of these gene trees were chosen by Bayesian information criterion (BIC): LG+G4 for OSC, P450-2, P450-3, and SDR; JTT+G4 for AT-1; and LG+I+G4 for P450-1. The types of fusidane BGCs were marked with different colors in the nodes. The trees were visualized using ggtree package in R, and were drawn to scale, with branch lengths measured in the number of substitutions per site. Numbers at nodes indicate the bootstrap support from 1,000 ultrafast bootstrap replicates from IQ-TREE.