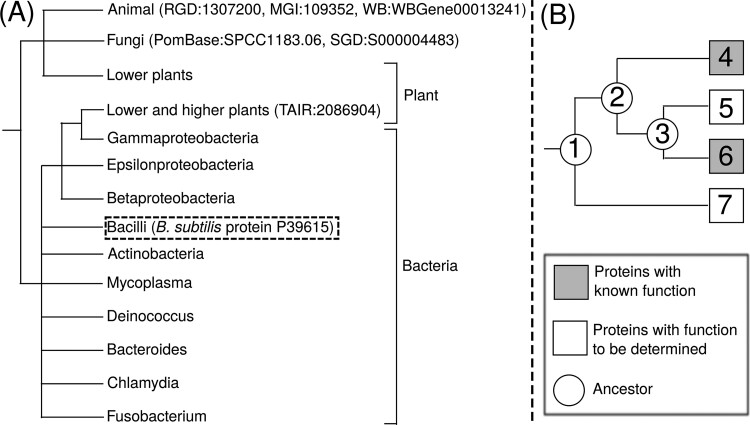
Fig. 3.
Phylogeny based annotation of IBA GO terms. (A) A simplified phylogenetic tree for Udg protein family (PTN000137400 in PANTHER database). The six orthologous proteins with experimentally annotated functions are shown in parentheses. The B.subtilis target protein to be annotated with IBA GO term is shown in parentheses with dashed box. The full phylogenetic tree for this PANTHER family is provided as Supplementary Figure S1 to S5. (B) A diagram for GO term annotation using a phylogenetic tree for a protein family with four member proteins (Squares 4–7), where two proteins (Squares 4 and 6 in grey) have the same experimentally annotated GO terms, while the function of the other two proteins (Squares 5 and 7 in white) are to be determined. Among the inferred biological ancestors (Circle 1–3) in this phylogenetic tree, ancestor 2 is the most recent common ancestor (MRCA) of Proteins 4 and 6. Upon manual inspection of the tree, curators often infer that the whole branch rooted by MRCA (Ancestor 2) share the same GO term as the leaf proteins (Protein 4–6), and assign the GO term annotation of Proteins 4 and 6 to Protein 5. The function of Protein 7, which belongs to an outgroup and does not have the same MRCA as 4 and 6, is usually left unassigned