Abstract
Long intergenic nonprotein-coding RNA 02163 (LINC02163) has been reported to be upregulated and work as an oncogene in gastric cancer. The aims of the present study were to determine the expression profile and clinical value of LINC02163 in breast cancer. Additionally, the detailed functions of LINC02163 in breast cancer were explored, and relevant molecular events were elucidated. In this study, LINC02163 was upregulated in breast cancer, and its expression level was closely associated with tumor size, lymph node metastasis, and TNM stage. Patients with breast cancer presenting high LINC02163 expression exhibited shorter overall survival than those presenting low LINC02163 expression. Knockdown of LINC02163 resulted in a decrease in breast cancer cell proliferation, migration, and invasion and an increase in cell apoptosis in vitro. In addition, silencing of LINC02163 impeded breast cancer tumor growth in vivo. Mechanistic investigation revealed that LINC02163 served as a competing endogenous RNA for microRNA-511-3p (miR-511-3p) and consequently upregulated the expression of the high-mobility group A2 (HMGA2), a downstream target of miR-511-3p. Intriguingly, miR-511-3p inhibition and HMGA2 restoration counteracted the effects of LINC02163 deficiency on the malignant properties of breast cancer cells. LINC02163 exerts cancer-promoting effects during the initiation and progression of breast cancer via regulation of the miR-511-3p/HMGA2 axis. Our findings add to our understanding of the roles of the LINC02163/miR-511-3p/HMGA2 pathway as a regulator of breast cancer pathogenesis and may be useful in the development of lncRNA-directed cancer diagnosis, prognosis, and therapy.
Key words: Long intergenic nonprotein-coding RNA 02163 (LINC02163), High-mobility group A2 (HMGA2), MicroRNA-511-3p (miR-511-3p), Breast cancer
INTRODUCTION
Breast cancer ranks as the second leading cause of cancer-associated death among women globally1, affecting approximately 1.7 million individuals and causing approximately 500,000 mortalities worldwide each year2. At present, the first-line therapies for breast cancer are surgical resection, radiotherapy, chemotherapy, endocrine therapy, and immunotherapy3. Dramatic improvements in diagnostic tools and therapeutic methods have markedly improved clinical outcomes for patients with breast cancer; nevertheless, the long-term survival of patients with advanced stage cancer remains unsatisfactory, primarily because of high rates of recurrence and distant metastasis4,5. The 5-year overall survival rate for early stage breast cancer is as high as 90%; unfortunately, it is approximately 6% in patients diagnosed at an advanced stage6. Although multiple mechanisms and pathways underlying the pathogenesis of breast cancer have been intensively explored7–9, the details of these mechanisms are still largely unknown and require further study. Therefore, it is essential to elucidate the mechanisms contributing to breast carcinogenesis and progression so as to identify effective targets for cancer diagnosis, prognosis, and management.
Long noncoding RNAs (lncRNAs) are a category of transcripts that are generally >200 nucleotides in length but lack extended open reading frames10. To date, >3,000 lncRNAs have been verified in the human genome; they are important gene regulators and play key roles in normal development, physiological processes, and human disease11–13. Vast amounts of concrete research have revealed that aberrant expression of lncRNAs is frequently observed in many human diseases, especially cancers14. Many lncRNAs are dysregulated in breast cancer, and this dysregulation is closely associated with the oncogenicity of the cancer15–17. It has been shown that lncRNAs drive the malignant behavior of breast cancer through cancer-inhibiting or cancer-promoting effects18,19.
MicroRNAs (miRNAs) belong to a subgroup of single-stranded, noncoding RNA molecules that are 17–25 nucleotides in length20. miRNAs are able to negatively regulate gene expression by directly binding to the 3′-untranslated region (3′-UTR) of targeted messenger RNA (mRNA), resulting in the degradation of target mRNAs and/or the suppression of translation21. Several studies have revealed that during tumor initiation and progression, lncRNAs operate as competing endogenous RNAs (ceRNAs) for miRNAs and consequently decrease the regulatory impacts of these miRNAs on their target mRNAs22–24. In view of this, understanding the detailed functions of lncRNAs and miRNAs that contribute to breast cancer malignancy as well as elucidating the mechanisms of their action may provide potential targets for anticancer management strategies for breast cancer.
The lncRNA long intergenic nonprotein-coding RNA 02163 (LINC02163) is recognized as a vital regulator of gastric cancer25. At present, it is largely unclear whether LINC02163 contributes to the malignancy of breast cancer. Accordingly, the aims of the present study were to determine the expression profile and clinical value of LINC02163 in breast cancer. Additionally, the functions of LINC02163 in breast cancer were explored in detail, and relevant molecular events were elucidated. The results of this study may vastly improve our understanding of the roles and molecular mechanisms of LINC02163 in the pathogenesis of breast cancer.
MATERIALS AND METHODS
Clinical Samples
The present study was performed with the approval of the ethics committees of Yancheng City No. 1 People’s Hospital (Yancheng, China) and conducted in accordance with the provisions of the Helsinki Declaration. Written informed consent was obtained from all enrolled participants. In total, 61 paired breast cancer tissues and matched adjacent normal tissues were collected from patients admitted to Yancheng City No. 1 People’s Hospital. None of the patients had previously undergone preoperative radiotherapy, chemotherapy, endocrine therapy, immunotherapy, or other anticancer treatments. After tumor excision, all tissue samples were immediately frozen and preserved in liquid nitrogen until further use.
Cell Culture
Human immortalized breast epithelial cell line MCF-10A was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in MEGM™ Mammary Epithelial Cell Growth Medium BulletKit™ (Lonza/Clonetics Corporation, Walkersville, MD, USA) containing 100 ng/ml of cholera toxin. Breast cancer cell lines SKBR3, MDA-MB-231, MCF-7, and BT-474 (all from ATCC) were cultured in McCoy’s 5a Medium (Gibco, Grand Island, NY, USA), Leibovitz’s L-15 medium (ATCC), Eagle’s minimum essential medium (ATCC), and Hybri-Care medium (ATCC), respectively, to which was added 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Gibco). All cells were maintained at 37°C in a humidified environment with 5% CO2.
Cell Transfection
Three LINC02163-specific small interfering RNAs (termed si-LINC02163#1, si-LINC02163#2, and si-LINC02163#3) and negative control (NC) small interfering RNA (termed si-NC) were designed and chemically synthesized by RiboBio Co, Ltd (Guangzhou, China). Additionally, miR-511-3p mimic, NC miRNA mimic (termed miR-NC), miR-511-3p inhibitor, and NC inhibitor were obtained from GenePharma (Shanghai, China). The high-mobility group A2 (HMGA2) overexpression plasmid pcDNA3.1-HMGA2 (termed pc-HMGA2) and an empty pcDNA3.1 plasmid were prepared by Shanghai GeneChem Co, Ltd (Shanghai, China). Breast cancer cells were seeded into six-well plates. Using Lipofectamine 2000™ reagent (Invitrogen; Thermo Fisher Scientific, Carlsbad, CA, USA), breast cancer cells were transiently transfected with the abovementioned siRNA (100 pmol), miRNA mimic (100 pmol), miRNA inhibitor (100 pmol), or plasmids (4 μg).
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific) was used to isolate total RNA from the cells. The concentration and purity of the RNA were evaluated using a spectrophotometer (Bio-Rad, Hercules, CA, USA). Total RNA was reverse transcribed into first-strand cDNA using the PrimeScript RT Reagent kit (TaKaRa, Dalian, China). Then quantitative PCR was performed to determine LINC02163 and HMGA2 mRNA expression using SYBR Premix Ex Taq (TaKaRa). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the normalization control for LINC02163 and HMGA2 mRNA.
To assess miR-511-3p expression, reverse transcription and quantitative PCR were performed using the miScript Reverse Transcription kit (Qiagen, Hilden, Germany) and the miScript SYBR Green PCR Kit (Qiagen), respectively. The expression of miR-511-3p was normalized to U6 small nuclear RNA. The 2−ΔΔCt method was employed for data analysis.
The primers were designed as follows: LINC02163, 5′- TGCATGCTGTTTAGAAGGCAG-3′ (forward) and 5′-TGCTACTGGTTGCAGGATTT-3′ (reverse); HMGA2, 5′-GGCTCAGATTCAGGAACAGC-3′ (forward) and 5′-GCTTCAACGGCAAAGTTCTC-3′ (reverse); GAPDH, 5′-GACAGTCAGCCGCATCTTCT-3′ (forward) and 5′-GCGCCCAATACGACCAAATC-3′ (reverse); miR-511-3p, 5′-ACACCCATCGTGTCTTTTGC-3′ (forward) and 5′-CAATGGACCACCATTCTGTCT-3′ (reverse); and U6, 5′-GCGCGTCGTGAAGCGTTC-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse).
Nuclear/Cytoplasmic Fractionation
The nuclear and cytoplasmic fractions of breast cancer cells were separated using a PARIS™ kit (Invitrogen) in accordance with the supplier’s specifications. GAPDH and U6 small nuclear RNA were used as the nuclear and cytoplasmic internal references, respectively.
Cell Counting Kit-8 (CCK-8) Assay
Transfected cells were harvested at 24-h posttransfection, and complete culture medium was mixed with the transfected cells to prepare a cell suspension. The concentration of the cell suspension was adjusted to 2 × 104 cells per ml. A volume of 100 μl of cell suspension was seeded into 96-well plates. Following cell inoculation, cell proliferation was evaluated using the CCK-8 assay once per day for 3 consecutive days. A total of 10 μl of CCK-8 solution (Sigma-Aldrich, St. Louis, MO, USA) was added to each well, followed by incubation for an additional 2 h at 37°C. The absorbance value of each well was detected at a wavelength of 450 nm using a microplate reader (NYW-96M; Beijing Nyaw Instrument & Meter Co, Ltd, Beijing, China).
Flow Cytometry
After 48 h of culture, transfected cells were removed from the culture plates using ethylenediaminetetraacetic acid-free 0.05% trypsin (Gibco) and rinsed twice with precooled phosphate buffer solution. The proportion of apoptotic cells was quantified using an Annexin-V–Fluorescein Isothiocyanate (FITC) Apoptosis Detection kit (Biolegend, San Diego, CA, USA). Following centrifugation, the supernatant was discarded, and transfected cells were resuspended in 200 μl of binding buffer; then 5 μl of FITC–Annexin-V and 10 μl of propidium iodide were added. After incubation for 15 min in darkness, data on cell apoptosis were acquired using flow cytometry (FACScan; BD Biosciences, Franklin Lakes, NJ, USA) and analyzed using Cell Quest acquisition software (version 2.9; BD Biosciences).
Transwell Cell Migration and Invasion Assays
To evaluate the migratory and invasive capacities of the cancer cells, 24-well Transwell inserts with an aperture of 8 μm were used. Matrigel (30 μg/well; BD Biosciences, San Jose, CA, USA) was prelaid in the top inserts prior to use in the Transwell invasion assay. The subsequent experimental procedures for migration and invasion assays were the same. Briefly, transfected cells were harvested, resuspended in FBS-free culture medium, and inoculated into the upper inserts. The bottom chambers were filled with culture medium containing 20% FBS. The chambers were maintained at 37°C for 24 h. Next, nonmigratory and noninvasive cells were gently removed with a cotton swab. The cells that passed through the pores in the membranes were fixed with 4% polyformaldehyde and stained with 0.1% crystal violet. After extensive rinsing, the migratory and invasive cells were observed and imaged under an inverted microscope (Olympus, Tokyo, Japan).
Xenograft Tumor Model
To construct MDA-MB-231 cells with stably depleted of LINC02163, short hairpin RNA (shRNA) against the expression of LINC02163 (sh-LINC02163) and NC shRNA (sh-NC; both from GenePharma) were inserted into the pLKO.1 vector (Biosettia, San Diego, CA, USA) to generate the plasmids pLKO.1-sh-LINC02163 and pLKO.1-sh-NC, respectively. The plasmids were separately transfected into MDA-MB-231 cells, which were then incubated with puromycin to select cells in which LINC02163 was stably silenced.
The animal study protocol was approved by the Institutional Animal Care and Use Ethics Committee of Yancheng City No 1 People’s Hospital (Yancheng, China). Female 4-week-old BALB/c nude mice (Shanghai Slac Experimental Animals Co, Ltd, Shanghai, China) were raised in a specific pathogen-free environment and subcutaneously injected with MDA-MB-231 cells stably expressing sh-LINC02163 or sh-NC. Each group contained three nude mice. One week after cell inoculation, the width and length of tumor xenografts were measured using a vernier caliper weekly. The tumor volumes were determined according to the following equation: tumor volume (mm3) = 0.5 × length (mm) × width2 (mm2). At week 5 postinoculation, all mice were euthanatized, and subcutaneous xenografts were taken from the euthanatized mice. After weighing the tumor xenografts, they were stored in liquid nitrogen for further use.
Bioinformatic Predictions
starBase 3.0 (http://starbase.sysu.edu.cn/) was employed to predict putative miRNAs able to bind to LINC02163. The potential targets of miR-511-3p were searched using Targetscan 7.2 (http://www.targetscan.org/) and miRDB (http://mirdb.org/).
Luciferase Reporter Assay
Fragments of LINC02163 carrying the wild-type (wt) or mutant (mut) miR-511-3p binding site were amplified by GenePharma and separately inserted into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation, Madison, WI, USA). The resultant luciferase reporter plasmids were designated as wt-LINC02163 and mut-LINC02163, respectively. Additionally, wt-HMGA2 and mut-HMGA2 reporter plasmids were synthesized using the same experimental procedures. For the reporter assay, the constructed reporter plasmids were cotransfected with miR-511-3p mimic or miR-NC into breast cancer cells. At 48 h posttransfection, the activities of firefly and Renilla luciferases were determined using a dual-luciferase reporter assay (Promega Corporation). The activity of firefly luciferase was normalized to that of Renilla luciferase.
RNA Immunoprecipitation (RIP) Assay
An RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore Sigma, Burlington, MA, USA). Breast cancer cells were collected and lysed in RIP buffer, and the whole-cell lysates obtained were incubated with RIP buffer containing magnetic beads conjugated with human Ago2 antibody (Millipore). Normal immunoglobulin (Ig) G (Millipore) served as the NC. The magnetic beads were collected after overnight incubation. The immunoprecipitated RNA was extracted and analyzed using RT-qPCR to assess whether LINC02163 and miR-511-3p were coimmunoprecipitated.
Western Blotting
Cells were lysed in RIPA lysis buffer supplemented with 1% phenylmethylsulfonyl fluoride (Beyotime, Shanghai, China). A BCA protein assay kit (Beyotime) was used to determine total protein concentrations. Equivalent proteins were separated on 10% gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride membranes. After blocking with 5% defatted milk powder, the membranes carrying the transferred proteins were incubated with primary antibodies against HMGA2 (dilution 1:1,000; ab207301; Abcam, Danvers, MA, USA) or GAPDH (dilution 1:1,000; ab181603; Abcam). Subsequently, immunodetection was performed using a goat anti-rabbit HRP-labeled secondary antibody (dilution 5:1,000; ab205718; Abcam) and Enhanced Chemiluminescence Reagent (Bio-Rad Laboratories). GAPDH was used as the loading control.
Statistical Analysis
All experiments were performed in triplicate and repeated at least three times. The results were presented as means ± standard deviation. SPSS software (version 18.0; SPSS, Inc, Chicago, IL, USA) was employed to process all data. Correlations between LINC02163 expression and clinicopathological features present in patients with breast cancer were evaluated using the chi-square test. The Student’s t-test was used for comparisons between two groups, and one-way analysis of variance followed by Tukey’s test was performed to compare differences among multiple groups. The correlation between LINC02163 and miR-511-3p expression was assessed using Spearman’s correlation analysis. The same method was also used to test the correlation between miR-511-3p and HMGA2 as well as that between LINC02163 and HMGA2 mRNA. The relationship between LINC02163 expression and the overall survival of patients with breast cancer was determined using the Kaplan–Meier method and a log-rank test. A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
Elevated LINC02163 Expression in Breast Cancer Is Associated With Poor Prognosis
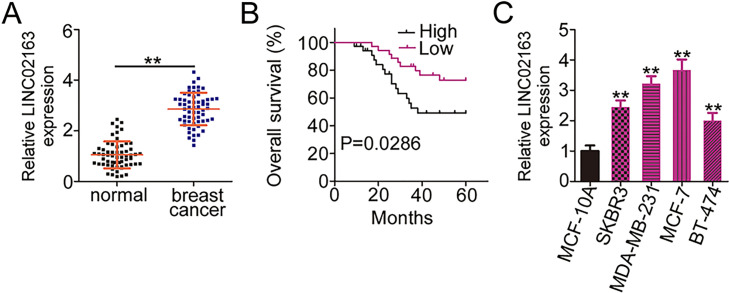
To determine whether LINC02163 is implicated in breast cancer pathogenesis, RT-qPCR was first performed to detect the expression of LINC02163 in 61 paired breast cancer tissues and matched adjacent normal tissues. The results showed that LINC02163 was markedly upregulated in breast cancer tissues compared with that in adjacent normal tissues (Fig. 1A). Next, we analyzed the clinical relevance of LINC02163 in patients with breast cancer. All patients with breast cancer were classified into either a low-LINC02163 (n = 30) group or a high-LINC02163 (n = 31) group based on the median amount of LINC02163 in their breast cancer tissues. Kaplan–Meier survival curves revealed that the patients in the high-LINC02163 group exhibited shorter overall survival than the patients in the low-LINC02163 group (p = 0.0286) (Fig. 1B). In addition, LINC02163 expression was strongly correlated with tumor size (p = 0.041), lymph node metastasis (p = 0.002), and TNM stage (p = 0.013) (Table 1). Furthermore, RT-qPCR analysis revealed that all four breast cancer cell lines (SKBR3, MDA-MB-231, MCF-7, and BT-474) expressed LINC02163 at relatively higher levels than that in human immortalized breast epithelial cell line MCF-10A (Fig. 1C). Together, the available evidence suggests that LINC02163 is highly expressed in breast cancer and may play cancerogenic roles in breast cancer.
Figure 1.
Long intergenic nonprotein-coding RNA 02163 (LINC02163) is upregulated in breast cancer and is correlated with poor prognosis. (A) Relative LINC02163 expression was detected in 61 pairs of breast cancer tissues and matched adjacent normal tissues using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). (B) The overall survival of patients with breast cancer was determined according to the Kaplan–Meier method, and differences between the two groups were examined using the log-rank test (p = 0.0286). (C) LINC02163 expression was measured using RT-qPCR in four breast cancer cell lines (SKBR3, MDA-MB-231, MCF-7, and BT-474) and the human immortalized breast epithelial cell line MCF-10A. **p < 0.01.
Table 1.
The Correlation of LINC02163 Level With Clinicopathological Features of Patients With Breast Cancer
| Clinicopathological Features | LINC02163 Expression | p Value | |
|---|---|---|---|
| High (31) | Low (30) | ||
| Age (years) | 0.446 | ||
| <60 | 18 | 14 | |
| ≥60 | 13 | 16 | |
| Tumor size (mm) | 0.041* | ||
| <2 | 12 | 20 | |
| ≥2 | 19 | 10 | |
| ER status | 0.800 | ||
| Negative | 14 | 15 | |
| Positive | 17 | 15 | |
| PR status | 0.611 | ||
| Negative | 15 | 17 | |
| Positive | 16 | 13 | |
| Lymph node metastasis | 0.002* | ||
| Negative | 10 | 22 | |
| Positive | 21 | 8 | |
| TNM stage | 0.013* | ||
| I–II | 16 | 25 | |
| III | 15 | 5 | |
| Differentiation grade | 0.340 | ||
| Well and moderately | 17 | 19 | |
| Poorly and undifferentiated | 14 | 11 | |
p < 0.05 by chi-square test.
LINC02163 Inhibition Impairs the Malignant Properties of Breast Cancer Cells In Vitro
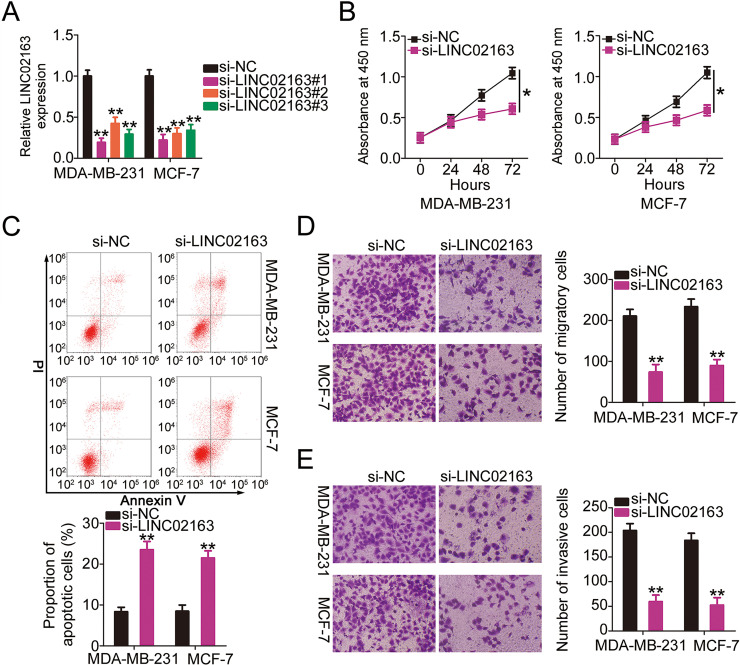
Given that LINC02163 expression was upregulated in breast cancer, a series of experiments was conducted to determine whether LINC02163 played a role in the oncogenicity of breast cancer. First, three siRNAs targeting LINC02163 were transfected into MDA-MB-231 and MCF-7 cell lines, and the interference efficiency was evaluated using RT-qPCR (Fig. 2A). The RT-qPCR data indicated that LINC02163 was silenced in varying degrees after transfection of the three siRNAs targeting LINC02163, among which si-LINC02163#1 presented the highest knockdown efficiency; this siRNA was renamed si-LINC02163 and was used in subsequent experiments. A CCK-8 assay showed that LINC02163 silencing markedly reduced the proliferation of MDA-MB-231 and MCF-7 cells compared with that of cells transfected with si-NC (Fig. 2B). Because of LINC02163 knockdown, MDA-MB-231 and MCF-7 cell apoptosis was markedly increased, as shown by flow cytometry (Fig. 2C). Furthermore, the impacts of LINC02163 downregulation on the metastatic capacity of breast cancer cells were investigated using Transwell cell migration and invasion assays. LINC02163 silencing resulted in an obvious reduction in the migratory (Fig. 2D) and invasive (Fig. 2E) abilities of MDA-MB-231 and MCF-7 cells. Collectively, these outcomes suggest that LINC02163 plays a pro-oncogenic role in the malignant properties of breast cancer cells in vitro.
Figure 2.
Impacts of LINC02163 silencing on the malignant properties of breast cancer cells in vitro. (A) Three siRNAs (si-LINC02163#1, si-LINC02163#2, and si-LINC02163#3) were designed to knock down endogenous LINC02163 expression in MDA-MB-231 and MCF-7 cells. RT-qPCR verified the transfection efficiency. (B) Proliferation was analyzed using the cell counting kit-8 (CCK-8) assay in MDA-MB-231 and MCF-7 cells after LINC02163 silencing. (C) Flow cytometry was conducted to determine the apoptosis rate of LINC02163-deficient MDA-MB-231 and MCF-7 cells. (D, E) The migration and invasion capacities of MDA-MB-231 and MCF-7 cells after LINC02163 silencing were evaluated using Transwell cell migration and invasion assays. *p < 0.05 and **p < 0.01.
LINC02163 Acts as a Molecular Sponge for miR-511-3p in Breast Cancer Cells
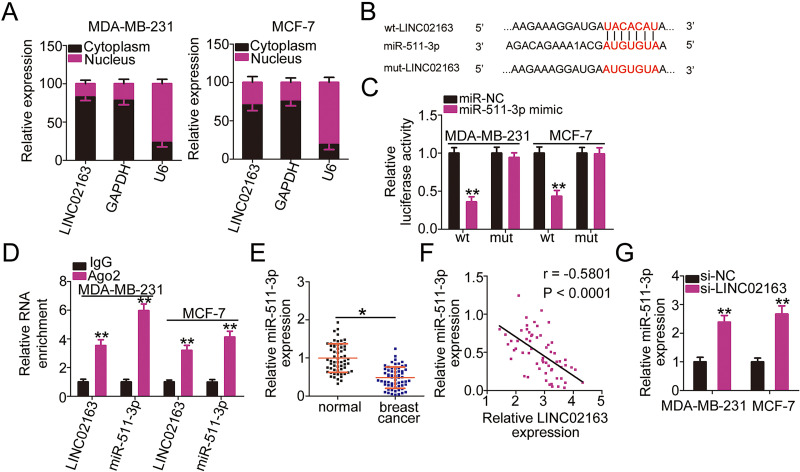
To elucidate the mechanism responsible for the cancer-promoting role of LINC02163, the expression distribution of LINC02163 in MDA-MB-231 and MCF-7 cells was first examined using nuclear/cytoplasmic fractionation. LINC02163 was mostly localized to the cytoplasm of MDA-MB-231 and MCF-7 cells (Fig. 3A), which suggests that LINC02163 serves as ceRNA or molecular sponges for specific miRNAs. Bioinformatic tools were employed to predict eligible miRNA that could directly bind to LINC02163. As shown in Figure 3B, miR-511-3p contained the putative complementary binding site for LINC02163. A luciferase reporter assay was conducted to confirm the functionality of this predicted binding site. After cotransfection with miR-511-3p mimic, wt-LINC02163-driven luciferase activity was strongly downregulated in MDA-MB-231 and MCF-7 cells, whereas the luciferase reporter expressing mut-LINC02163 was unaffected by miR-511-3p overexpression (Fig. 3C). Additionally, RIP assay indicated that LINC02163 and miR-511-3p were distinctly enriched in Ago2-containing beads compared with the IgG control group (Fig. 3D), indicating that miR-511-3p is capable of binding directly to LINC02163.
Figure 3.
LINC02163 functions as a molecular sponge for microRNA-511-3p (miR-511-3p) in breast cancer cells. (A) Nuclear/cytoplasmic fractionation coupled with RT-qPCR was conducted to assess the localization of LINC02163 expression in MDA-MB-231 and MCF-7 cells. (B) Predicted miR-511-3p binding site in LINC02163. Mutated binding sequences are also shown. (C) Luciferase reporter plasmids carrying wild-type or mutant miR-511-3p binding sites were cotransfected with miR-511-3p mimic or miR-NC into MDA-MB-231 and MCF-7 cells. Luciferase activity was detected using dual-luciferase reporter assay. (D) Enrichment of LINC02163 and miR-511-3p in MDA-MB-231 and MCF-7 cells was tested by RNA immunoprecipitation (RIP) assay. (E) Analysis of miR-511-3p expression in 61 pairs of breast cancer tissues and matched adjacent normal tissues was performed using RT-qPCR. (F) The correlation between LINC02163 and miR-511-3p expression levels in the 61 breast cancer tissues was examined using Spearman’s correlation analysis (r = −0.5801, p < 0.0001). (G) RT-qPCR was performed to measure miR-511-3p expression in MDA-MB-231 and MCF-7 cells when transfected with si-LINC02163 or si-NC. *p < 0.05 and **p < 0.01.
Furthermore, miR-511-3p was weakly expressed in breast cancer tissues compared with adjacent normal tissues (Fig. 3E). LINC02163 was inversely correlated with miR-511-3p in breast cancer tissues (r = −0.5801, p < 0.0001) (Fig. 3F), as evidenced by Spearman’s correlation analysis. RT-qPCR, performed to assess whether LINC02163 could regulate miR-511-3p expression in breast cancer cells, showed that transfection with si-LINC02163 caused an apparent upregulation of miR-511-3p expression in MDA-MB-231 and MCF-7 cells (Fig. 3G). In short, LINC02163 acted as a molecular sponge for miR-511-3p in breast cancer cells.
miR-511-3p Exerts Tumor-Suppressing Effects in Breast Cancer Cells via Direct Targeting of HMGA2
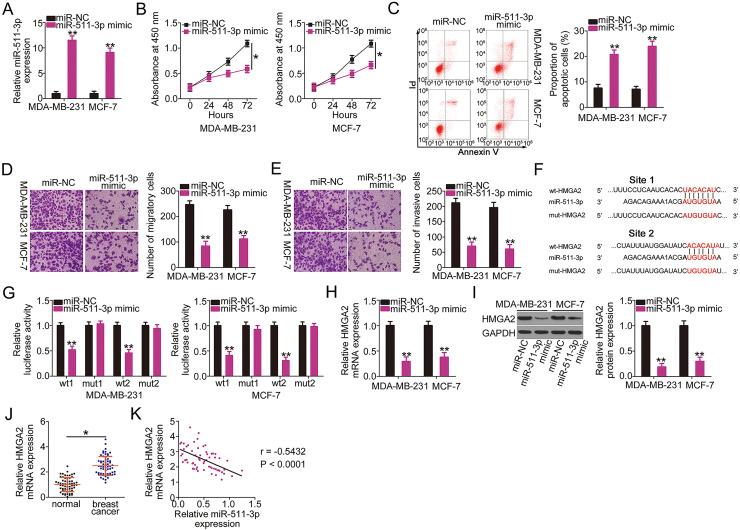
Next, the detailed roles of miR-511-3p in breast cancer progression were explored. The transfection efficiency of miR-511-3p mimic was tested by RT-qPCR, and the results showed that the miR-511-3p mimic was effective (Fig. 4A). A CCK-8 assay and flow cytometry were performed to assess the proliferation and apoptosis of breast cancer cells; the results revealed that exogenous miR-511-3p expression markedly suppressed MDA-MB-231 and MCF-7 cell proliferation (Fig. 4B) and promoted cell apoptosis (Fig. 4C). Transwell cell migration and invasion assays were also conducted to investigate the effects of miR-511-3p overexpression on breast cancer cell migration and invasion. The number of migratory (Fig. 4D) and invasive (Fig. 4E) cells decreased in MDA-MB-231 and MCF-7 cells treated with the miR-511-3p mimic.
Figure 4.
miR-511-3p acts as a tumor-suppressive miRNA in breast cancer and directly targets high-mobility group A2 (HMGA2). (A) The efficiency of miR-511-3p mimic transfection in MDA-MB-231 and MCF-7 cells was determined by RT-qPCR. (B, C) The effects of miR-511-3p upregulation on MDA-MB-231 and MCF-7 cell proliferation and apoptosis were evaluated using CCK-8 assay and flow cytometry. (D, E) The migration and invasion capabilities of MDA-MB-231 and MCF-7 cells with miR-511-3p overexpression were detected using Transwell cell migration and invasion assays. (F) The wild-type binding site exists between miR-511-3p and HMGA2. Mutant binding sequences are also shown. (G) Luciferase activity was detected in MDA-MB-231 and MCF-7 cells after cotransfection with wt-HMGA2 or mut-HMGA2 reporter plasmids and miR-511-3p mimic or miR-NC. (H, I) HMGA2 mRNA and protein expression in miR-511-3p mimic-transfected or miR-NC-transfected MDA-MB-231 and MCF-7 cells were examined by RT-qPCR and Western blotting, respectively. (J) HMGA2 mRNA expression was examined in 61 pairs of breast cancer tissues and matched adjacent normal tissues using RT-qPCR. (K) Spearman’s correlation analysis was applied to dissect the expression correlation between miR-511-3p and HMGA2 mRNA in the 61 breast cancer tissues (r = −0.5432, p < 0.0001). *p < 0.05 and **p < 0.01.
Bioinformatic predictions using data from online miRNA target prediction databases showed that the 3′-UTR of HMGA2 possesses two putative binding sites for miR-511-3p (Fig. 4F). Binding between miR-511-3p and HMGA2 was confirmed by the luciferase reporter assay. The luciferase activity of wt-HMGA2 (both 1 and 2) was strongly restricted in MDA-MB-231 and MCF-7 cells by miR-511-3p overexpression; nevertheless, the mutant form (both 1 and 2) with disruption of the potential miR-511-3p binding sequences was unaltered by miR-511-3p mimic cotransfection (Fig. 4G). To further investigate the direct regulation of HMGA2 by miR-511-3p, the mRNA and protein levels of HMGA2 in miR-511-3p-overexpressing MDA-MB-231 and MCF-7 cells were measured by RT-qPCR and Western blotting, respectively. HMGA2 expression at the mRNA (Fig. 4H) and protein (Fig. 4I) level in both cell types was downregulated by miR-511-3p mimic. Furthermore, the expression profile of HMGA2 was determined in tumor tissues and matched normal tissue samples from 61 patients with breast cancer. The expression of HMGA2 mRNA was upregulated in breast cancer tissues compared with that in adjacent normal tissues (Fig. 4J). Spearman’s correlation analysis identified an inverse correlation between miR-511-3p and HMGA2 mRNA in the 61 breast cancer tissues (r = −0.5432, p < 0.0001) (Fig. 4K). Collectively, these results indicate that miR-511-3p functions as a tumor-suppressing miRNA in breast cancer cells and that HMGA2 is a direct target of miR-511-3p.
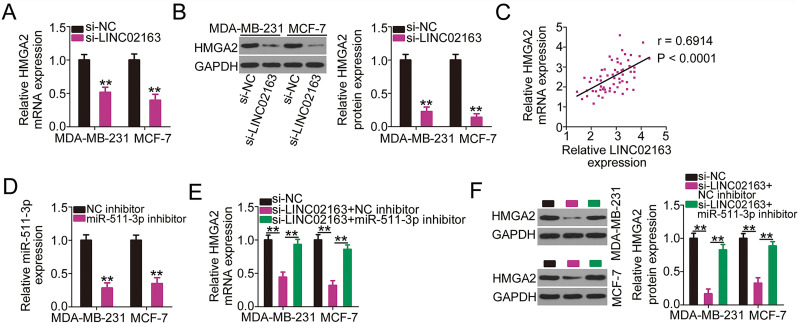
LINC02163 Increased HMGA2 Expression in Breast Cancer Cells Through Sponging miR-511-3p
As LINC02163 acts as a sponge for miR-511-3p and HMGA2 is a target of the latter, we next examined whether LINC02163 is implicated in the control of HMGA2 in breast cancer cells. RT-qPCR and Western blotting were used to detect HMGA2 mRNA and protein expression in MDA-MB-231 and MCF-7 cells after LINC02163 downregulation. Suppressing LINC02163 expression considerably restricted the expression of HMGA2 in MDA-MB-231 and MCF-7 cells at both the mRNA (Fig. 5A) and protein (Fig. 5B) levels. The correlation between LINC02163 and HMGA2 was examined in breast cancer tissues. As expected, HMGA2 mRNA was positively correlated with LINC02163 expression in the 61 breast cancer tissue samples (r = 0.6914, p < 0.0001) (Fig. 5C). To examine whether LINC02163 positively modulated HMGA2 expression by sponging miR-511-3p, rescue experiments were performed. First, RT-qPCR analysis confirmed the efficiency of miR-511-3p inhibitor in MDA-MB-231 and MCF-7 cells (Fig. 5D). Then si-LINC02163 was introduced in parallel with miR-511-3p inhibitor or NC inhibitor into MDA-MB-231 and MCF-7 cells, and the cotransfected cells were subsequently analyzed by RT-qPCR and Western blotting to determine the change in HMGA2 expression. The results showed that LINC02163 silencing reduced the mRNA (Fig. 5E) and protein (Fig. 5F) levels of HMGA2, and miR-511-3p inhibitor cotransfection abolished this decrease in HMGA2 expression triggered by LINC02163 knockdown. Thus, LINC02163 functions as a ceRNA for miR-511-3p in breast cancer and thereby increases HMGA2 expression.
Figure 5.
LINC02163 knockdown decreases HMGA2 expression in breast cancer cells through sponging miR-511-3p. (A, B) The mRNA and protein levels of HMGA2 in LINC02163-depleted MDA-MB-231 and MCF-7 cells were analyzed using RT-qPCR and Western blotting, respectively. (C) Spearman’s correlation analysis was performed to examine the correlation between LINC02163 and HMGA2 mRNA levels in 61 breast cancer tissues (r = 0.6914, p < 0.0001). (D) The knockdown efficiency of miR-511-3p inhibitor in MDA-MB-231 and MCF-7 cells was detected using RT-qPCR. (E, F) si-LINC02163 along with miR-511-3p inhibitor or negative control (NC) inhibitor was cotransfected into MDA-MB-231 and MCF-7 cells. The mRNA and protein levels of HMGA2 were detected using RT-qPCR and Western blotting, respectively. **p < 0.01.
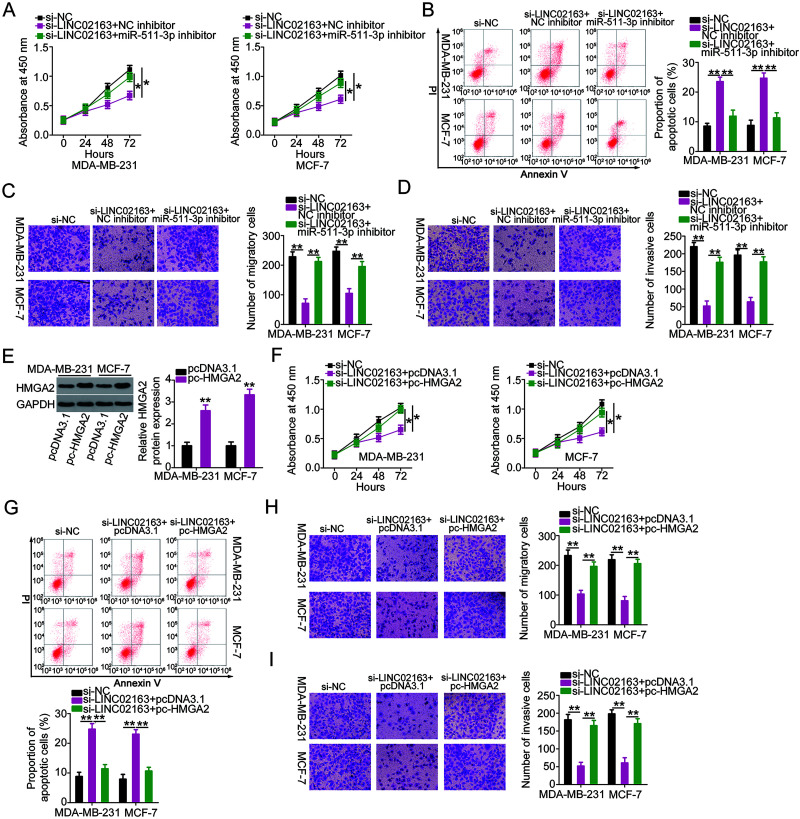
The miR-511-3p/HMGA2 Axis Is Involved in the Pro-Oncogenic Activities of LINC02163 in Breast Cancer Cells
Rescue experiments were conducted with the aim of further confirming that the actions of LINC02163 in breast cancer cells are mediated by its influences on the miR-511-3p/HMGA2 axis. First, MDA-MB-231 and MCF-7 cells were transfected with miR-511-3p inhibitor or NC inhibitor in the presence of si-LINC02163. A CCK-8 assay and flow cytometry revealed that suppression of miR-511-3p rescued the inhibition of MDA-MB-231 and MCF-7 cell proliferation (Fig. 6A) and the induction of cell apoptosis (Fig. 6B), which were a result of LINC02163 depletion. Simultaneously, Transwell cell migration and invasion assays verified that LINC02163 knockdown hindered the migratory (Fig. 6C) and invasive (Fig. 6D) abilities of MDA-MB-231 and MCF-7 cells, but these impacts were attenuated in response to miR-511-3p inhibitor cotransfection. Rescue experiments were also designed and performed with MDA-MB-231 and MCF-7 cells by cotransfecting with si-LINC02163 and pc-HMGA2 or pcDNA3.1. Western blotting confirmed that transfection with pc-HMGA2 considerably increased the expression of HMGA2 protein in both cell types (Fig. 6E). Recovery of HMGA2 expression neutralized the effects of LINC02163 on proliferation (Fig. 6F), apoptosis (Fig. 6G), migration (Fig. 6H), and invasion (Fig. 6I) in MDA-MB-231 and MCF-7 cells. Collectively, these results suggest that LINC02163 exerts its oncogenic actions during breast cancer progression by increasing the output of the miR-511-3p/HMGA2 axis.
Figure 6.
Elevation output of the miR-511-3p/HMGA2 axis abrogates the effects of LINC02163 knockdown in the malignant properties of breast cancer cells. (A, B) miR-511-3p inhibitor or NC inhibitor was introduced to MDA-MB-231 and MCF-7 cells in the presence of si-LINC02163. CCK-8 assay and flow cytometry were used to examine cell proliferation and apoptosis. (C, D) Migration and invasion of MDA-MB-231 and MCF-7 cells treated as described above were analyzed using Transwell cell migration and invasion assays. (E) The protein level of HMGA2 in MDA-MB-231 and MCF-7 cells following transfection of pc-HMGA2 or pcDNA3.1 was measured using Western blotting. (F–I) si-LINC02163 in combination with pc-HMGA2 or pcDNA3.1 was transfected into MDA-MB-231 and MCF-7 cells. After transfection, cell proliferation, apoptosis, migration, and invasion were analyzed using CCK-8 assay, flow cytometry, and Transwell cell migration and invasion assays, respectively. *p < 0.05 and **p < 0.01.
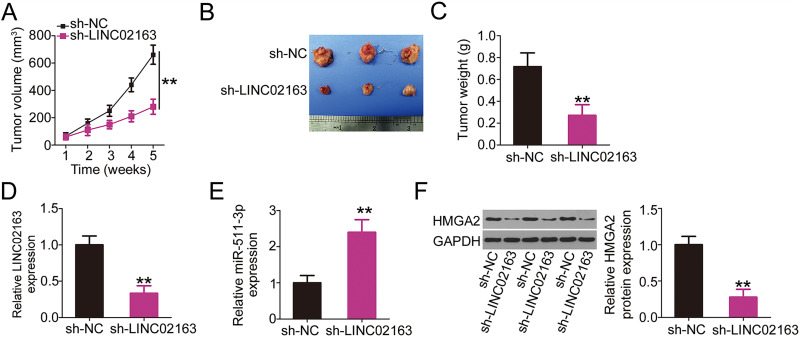
LINC02163 Downregulation Suppresses Breast Cancer Tumor Growth In Vivo
To investigate the effect of LINC02163 on breast cancer tumor growth in vivo, a xenograft mouse model was established by subcutaneously injecting MDA-MB-231 cells stably expressing sh-LINC02163 or sh-NC into mice. Tumor growth was considerably slowed in the sh-LINC02163 group compared with that in the sh-NC group (Fig. 7A). Five weeks after cell injection, subcutaneous xenografts were harvested and photographed (Fig. 7B). The weight of the subcutaneous xenografts originating from LINC02163-deficient MDA-MB-231 cells was significantly lower than that of xenografts originating from cells expressing sh-NC (Fig. 7C). Additionally, LINC02163, miR-511-3p, and HMGA2 expression in the subcutaneous xenografts was determined. RT-qPCR analysis revealed a significant reduction in LINC02163 expression in the subcutaneous xenografts harvested from the sh-LINC02163 group (Fig. 7D). Furthermore, miR-511-3p upregulation (Fig. 7E) and HMHA2 protein (Fig. 7F) downregulation were observed in the tumor xenografts originating from sh-LINC02163 transfection, which was in accordance with the results of in vitro experiments. Collectively, these results indicate that LINC02163 silencing restricted breast cancer tumor growth in vivo, presumably through the miR-511-3p/HMGA2 axis.
Figure 7.
A reduction in LINC02163 expression attenuates breast tumor growth in vivo. (A) Growth curve of subcutaneous xenografts harvested from sh-LINC02163 and sh-NC groups. (B) Representative images of subcutaneous xenografts originating from MDA-MB-231 cells stably expressing sh-LINC02163 or sh-NC. (C) All mice were anesthetized 5 weeks after cell injection. Tumor xenografts were resected and weighed. (D, E) LINC02163 and miR-511-3p levels in the tumor xenografts removed from nude mice were detected using RT-qPCR analysis. (F) The protein expression of HMGA2 in the tumor xenografts was determined using Western blotting. **p < 0.01.
DISCUSSION
Extensive efforts have contributed to the study of the mechanisms underlying breast carcinogenesis and cancer progression, which has aroused more and more concern26–28. A considerable number of lncRNAs are aberrantly expressed in breast cancer and are involved in the modulation of numerous biological activities29–31. Considering the importance of lncRNAs in breast cancer, these polynucleotides may be effective targets for breast cancer diagnosis, therapy, and prognosis. In the present study, we detected the expression of LINC02163 in breast cancer and assessed the clinical value of LINC02163 in patients with breast cancer. Additionally, we investigated the role of LINC02163 in the biological behavior of breast cancer cells in vitro and in vivo. Furthermore, the molecular mechanisms responsible for the LINC021633-dependent malignant characteristics of breast cancer cells were elucidated in detail.
LINC02163 is upregulated in gastric cancer, presenting a close relationship with tumor size, distant metastasis, and clinical stage25. In addition, patients with gastric cancer showing high LINC02163 expression exhibit shorter overall survival and lower disease-free survival rates than patients showing low LINC02163 expression25. Functionally, LINC02163 knockdown suppresses gastric cancer cell proliferation, colony formation, invasion, and epithelial–mesenchymal transition in vitro, promotes G0/G1 cell cycle arrest, and decreases tumor growth in vivo25. Nevertheless, the involvement of LINC02163 in breast cancer is far from well characterized. In the present study, LINC02163 was shown to be upregulated in both breast cancer tissues and breast cancer cell lines, and was correlated with tumor size, lymph node metastasis, TNM stage, and shorter overall survival. To further investigate whether LINC02163 drives the oncogenicity of breast cancer, we explored the influences of LINC02163 knockdown through a series of in vitro functional assays as well as in vivo experiments in a xenograft tumor model. The results revealed that silencing LINC02163 expression restricted cell proliferation, migration, and invasion in vitro; facilitated cell apoptosis; and suppressed tumor growth in vivo.
In the present study, the regulatory mechanisms of action of LINC02163 in breast cancer were also explored. As lncRNAs exert their biological functions during carcinogenesis and cancer progression through multiple mechanisms, mostly according to subcellular localization32, the expression distribution of LINC02163 was analyzed. LINC02163 was found to be predominately distributed in the cytoplasm of breast cancer cells. A great deal of research has confirmed that cytoplasmic lncRNAs can act as miRNA sponges to competitively interact with certain miRNAs and consequently relieve the inhibitory effects of miRNAs on their target mRNAs; this is the mechanism of ceRNAs33–35.
According to bioinformatic predictions, miR-511-3p was screened for further experimental identification as it has been shown to exhibit antioncogenic activities in multiple human cancer types36,37. In addition, the results of luciferase reporter and RIP assays verified that LINC02163 can bind directly and interact with miR-511-3p in breast cancer cells. An inverse correlation between LINC02163 and miR-511-3p expression was identified in breast cancer tissues. LINC02163 knockdown resulted in a significant increase in miR-511-3p and a decrease in HMGA2, a target of miR-511-3p, in breast cancer cells. Furthermore, rescue experiments indicated that LINC02163 exerts its positive regulatory effects on HMGA2 expression via sponging miR-511-3p. In summary, LINC02163, miR-511-3p, and HMGA2 form an interactive ceRNA pathway in breast cancer.
It has been shown that miR-511-3p exerts tumor-suppressing effects and is downregulated in lung adenocarcinoma36 and prostate cancer37. Thus far, no relevant studies have addressed the expression and detailed functions of miR-511-3p in breast cancer. In the present study, using a series of functional experiments, we found that miR-511-3p was weakly expressed in breast cancer and that exogenous miR-511-3p expression restricted the malignant phenotypes of breast cancer cells. Furthermore, mechanistic investigation validated HMGA2 as a direct target of miR-511-3p in breast cancer. HMGA2, a member of the high-mobility group A protein family, is highly expressed in breast cancer and plays cancer-promoting roles in the malignancy of breast cancer by affecting a wide range of pathological behaviors38–41. The results of the present study clearly indicate that expression of HMGA2 is directly regulated by the LINC02163/miR-511-3p axis in breast cancer. Rescue assays showed that the miR-511-3p/HMGA2 axis is necessary for LINC02163-mediated breast cancer progression. Therefore, LINC02163 drives the oncogenicity of breast cancer via the miR-511-3p/HMGA2 axis, indicating that the LINC02163/miR-511-3p/HMGA2 pathway is a novel potential diagnostic and prognostic biomarker as well as a therapeutic target for breast cancer.
In summary, LINC02163 exerts cancer-promoting effects during the initiation and progression of breast cancer. Mechanically, LINC02163 acts as a ceRNA for miR-511-3p and consequently increases HMGA2 expression. Our findings improve our understanding of the roles of the LINC02163/miR-511-3p/HMGA2 pathway as a regulator of breast cancer pathogenesis and may be useful in the development of lncRNA-directed diagnosis, prognosis, and therapy.
ACKNOWLEDGMENTS
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, Soria JC, Dien AT, Adnani Y, Kamal M, Garnier S, Meurice G, Jimenez M, Dogan S, Verret B, Chaffanet M, Bachelot T, Campone M, Lefeuvre C, Bonnefoi H, Dalenc F, Jacquet A, De Filippo MR, Babbar N, Birnbaum D, Filleron T, Le Tourneau C, Andre F. Genomic characterization of metastatic breast cancers. Nature 2019;569:560–4. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 3. Nagini S. Breast cancer: Current molecular therapeutic targets and new players. Anticancer Agents Med Chem. 2017;17:152–63. [DOI] [PubMed] [Google Scholar]
- 4. Lu J, Steeg PS, Price JE, Krishnamurthy S, Mani SA, Reuben J, Cristofanilli M, Dontu G, Bidaut L, Valero V, Hortobagyi GN, Yu D. Breast cancer metastasis: Challenges and opportunities. Cancer Res. 2009;69:4951–3. [DOI] [PubMed] [Google Scholar]
- 5. Hu X, Huang W, Fan M. Emerging therapies for breast cancer. J Hematol Oncol. 2017;10:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castrellon AB. Novel strategies to improve the endocrine therapy of breast cancer. Oncol Rev. 2017;11:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017;389:2430–42. [DOI] [PubMed] [Google Scholar]
- 8. Barnard ME, Boeke CE, Tamimi RM. Established breast cancer risk factors and risk of intrinsic tumor subtypes. Biochim Biophys Acta 2015;1856:73–85. [DOI] [PubMed] [Google Scholar]
- 9. Rojas K, Stuckey A. Breast cancer epidemiology and risk factors. Clin Obstet Gynecol. 2016;59:651–72. [DOI] [PubMed] [Google Scholar]
- 10. Akhade VS, Pal D, Kanduri C. Long noncoding RNA: Genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74. [DOI] [PubMed] [Google Scholar]
- 11. Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: The dark matter matters. Curr Opin Genet Dev. 2018;48:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. [DOI] [PubMed] [Google Scholar]
- 13. Rogoyski OM, Pueyo JI, Couso JP, Newbury SF. Functions of long non-coding RNAs in human disease and their conservation in Drosophila development. Biochem Soc Trans. 2017;45:895–904. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y, Tao Y, Liao Q. Long noncoding RNA: A crosslink in biological regulatory network. Brief Bioinform. 2018;19:930–45. [DOI] [PubMed] [Google Scholar]
- 15. Li RH, Chen M, Liu J, Shao CC, Guo CP, Wei XL, Li YC, Huang WH, Zhang GJ. Long noncoding RNA ATB promotes the epithelial-mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018;9:1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lv P, Qiu X, Gu Y, Yang X, Xu X, Yang Y. Long non-coding RNA SNHG6 enhances cell proliferation, migration and invasion by regulating miR-26a-5p/MAPK6 in breast cancer. Biomed Pharmacother. 2019;110:294–301. [DOI] [PubMed] [Google Scholar]
- 17. Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, Qin Q, Zhao L, Huang Q, Luo Z, Huang S, Wei Z. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Y, Shao A, Wang L, Hu K, Yu C, Pan C, Zhang S. The role of lncRNAs in the distant metastasis of breast cancer. Front Oncol. 2019;9:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Youness RA, Gad MZ. Long non-coding RNAs: Functional regulatory players in breast cancer. Noncoding RNA Res. 2019;4:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tufekci KU, Oner MG, Meuwissen RL, Genc S. The role of microRNAs in human diseases. Methods Mol Biol. 2014;1107:33–50. [DOI] [PubMed] [Google Scholar]
- 21. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 22. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 2019;234:10080–100. [DOI] [PubMed] [Google Scholar]
- 23. Yu Y, Gao F, He Q, Li G, Ding G. lncRNA UCA1 functions as a ceRNA to promote prostate cancer progression via sponging miR143. Mol Ther Nucleic Acids 2019;19:751–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu G, Li S, Liu P, Shi Y, Liu Y, Yang Z, Fan Z, Zhu W. LncRNA TUG1 functions as a ceRNA for miR-6321 to promote endothelial progenitor cell migration and differentiation. Exp Cell Res. 2020;388:111839. [DOI] [PubMed] [Google Scholar]
- 25. Dong L, Hong H, Chen X, Huang Z, Wu W, Wu F. LINC02163 regulates growth and epithelial-to-mesenchymal transition phenotype via miR-593-3p/FOXK1 axis in gastric cancer cells. Artif Cells Nanomed Biotechnol. 2018;46:607–15. [DOI] [PubMed] [Google Scholar]
- 26. Pawlowska E, Szczepanska J, Blasiak J. The long noncoding RNA HOTAIR in breast cancer: Does autophagy play a role? Int J Mol Sci. 2017;18:2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Campos-Parra AD, Lopez-Urrutia E, Orozco Moreno LT, Lopez-Camarillo C, Meza-Menchaca T, Figueroa Gonzalez G, Bustamante Montes LP, Perez-Plasencia C. Long non-coding RNAs as new master regulators of resistance to systemic treatments in breast cancer. Int J Mol Sci. 2018;19:2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pecero ML, Salvador-Bofill J, Molina-Pinelo S. Long non-coding RNAs as monitoring tools and therapeutic targets in breast cancer. Cell Oncol. (Dordr) 2019;42:1–12. [DOI] [PubMed] [Google Scholar]
- 29. Zheng P, Dong L, Zhang B, Dai J, Zhang Y, Wang Y, Qin S. Long noncoding RNA CASC2 promotes paclitaxel resistance in breast cancer through regulation of miR-18a-5p/CDK19. Histochem Cell Biol. 2019;152:281–91. [DOI] [PubMed] [Google Scholar]
- 30. Li X, Deng S, Pang X, Song Y, Luo S, Jin L, Pan Y. LncRNA NEAT1 silenced miR-133b promotes migration and invasion of breast cancer cells. Int J Mol Sci. 2019;20:3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng A, Song X, Zhang L, Zhao L, Mao X, Wei M, Jin F. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/beta-catenin pathway. J Exp Clin Cancer Res. 2019;38:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun F, Liang W, Tang K, Hong M, Qian J. Profiling the lncRNA-miRNA-mRNA ceRNA network to reveal potential crosstalk between inflammatory bowel disease and colorectal cancer. Peer J. 2019;7:e7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao H, Hu GM, Wang WL, Wang ZH, Fang Y, Liu YL. LncRNA TDRG1 functions as an oncogene in cervical cancer through sponging miR-330-5p to modulate ELK1 expression. Eur Rev Med Pharmacol Sci. 2019;23:7295–306. [DOI] [PubMed] [Google Scholar]
- 34. Wang M, Hu H, Wang Y, Huang Q, Huang R, Chen Y, Ma T, Qiao T, Zhang Q, Wu H, Chen Q, Han D, Wang G, Wang X. Long non-coding RNA TUG1 mediates 5-fluorouracil resistance by acting as a ceRNA of miR-197-3p in colorectal cancer. J Cancer 2019;10:4603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao L, Hu K, Cao J, Wang P, Li J, Zeng K, He X, Tu PF, Tong T, Han L. lncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence. Aging (Albany NY) 2019;11:7098–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li J, Ge J, Yang Y, Liu B, Zheng M, Shi R. Long noncoding RNA ZFPM2-AS1 is involved in lung adenocarcinoma via miR-511-3p/AFF4 pathway. J Cell Biochem. 2020;121:2534–42. [DOI] [PubMed] [Google Scholar]
- 37. Zhang F, Wu Z. Significantly altered expression of miR-511-3p and its target AKT3 has negative prognostic value in human prostate cancer. Biochimie 2017;140:66–72. [DOI] [PubMed] [Google Scholar]
- 38. Mansoori B, Mohammadi A, Asadzadeh Z, Shirjang S, Minouei M, Abedi Gaballu F, Shajari N, Kazemi T, Gjerstorff MF, Duijf PHG, Baradaran B. HMGA2 and Bach-1 cooperate to promote breast cancer cell malignancy. J Cell Physiol. 2019;234:17714–26. [DOI] [PubMed] [Google Scholar]
- 39. Wang MJ, Zhang H, Li J, Zhao HD. microRNA-98 inhibits the proliferation, invasion, migration and promotes apoptosis of breast cancer cells by binding to HMGA2. Biosci Rep. 2018;38:BSR20180571. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40. Sgarra R, Pegoraro S, Ros G, Penzo C, Chiefari E, Foti D, Brunetti A, Manfioletti G. High mobility group A (HMGA) proteins: Molecular instigators of breast cancer onset and progression. Biochim Biophys Acta Rev Cancer 2018;1869:216–29. [DOI] [PubMed] [Google Scholar]
- 41. Wu J, Zhang S, Shan J, Hu Z, Liu X, Chen L, Ren X, Yao L, Sheng H, Li L, Ann D, Yen Y, Wang J, Wang X. Elevated HMGA2 expression is associated with cancer aggressiveness and predicts poor outcome in breast cancer. Cancer Lett. 2016;376:284–92. [DOI] [PubMed] [Google Scholar]