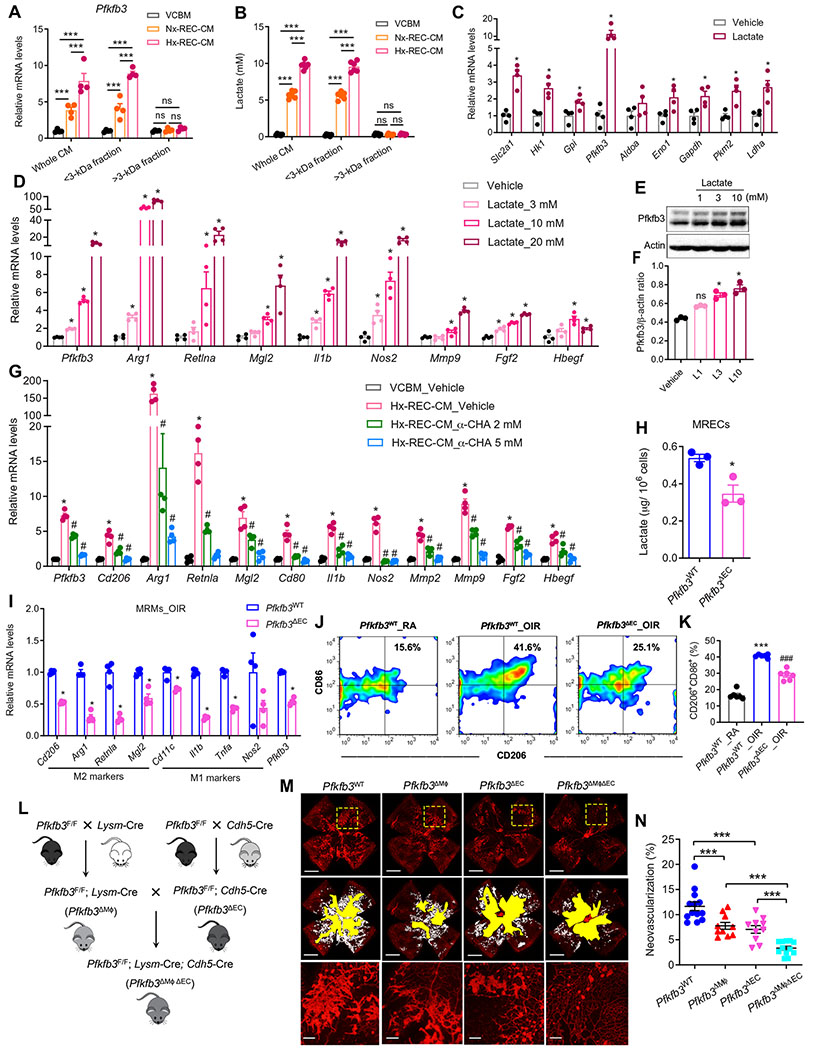
Fig. 7. REC-secreting lactate is sufficient to induce macrophage glycolysis and activation.
(A) VCBM, Nx-REC-CM, or Hx-REC-CM were used unfractionated (whole) or as >3-or <3-kDa fractions to stimulate BMDMs for 12 hours as follows. qRT-PCR analysis of the Mrna expression of Pfkfb3 in BMDMs. n = 4. ***P < 0.001 versus VCBM. ns, not significant. (B) Lactate concentration in medium from (A). n = 6. ***P < 0.001. (C) qRT-PCR analysis of the mRNA expression of glycolytic genes in BMDMs treated with 10 mM lactate for 12 hours (n = 4). *P < 0.05 versus vehicle. (D) qRT-PCR analysis of gene expression in BMDMs stimulated with a concentration gradient (3 to 20 mM) of lactate for 12 hours. n = 4. *P < 0.05 versus vehicle. (E and F) Western blot analysis and quantification of Pfkfb3 protein expression in BMDMs stimulated with lactate for indicated concentrations for 12 hours. n = 3. *P < 0.05 versus vehicle. (G) qRT-PCR analysis of gene expression in BMDMs exposed to VCBM or Hx-REC-CM for 12 hours, with or without α-cyano-4-hydroxycinnamic acid (α-CHA) (2 to 5 mM) treatment (n = 4). *P < 0.05 versus VCBM_Vehicle; #P < 0.05 versus Hx-REC-CM_Vehicle. (H) The amount of intracellular lactate of retinal ECs (MRECs) isolated from OIR Pfkfb3WT or Pfkfb3ΔMϕ mice at P17. *P < 0.05 versus Pfkfb3WT. (I) qRT-PCR analysis of gene expression in mouse retinal macrophages/microglia isolated from OIR Pfkfb3WT or Pfkfb3ΔEC mice at P17 (n = 4). *P < 0.05 versus Pfkfb3WT. (J and K) Representative flow cytometric plots of retinal CD11b+F4/80+ cells showed CD86+CD206+ fractions in the retinas of RA and OIR mice at P17 (J) and comparisons of CD86+CD206+ fractions between genotypes (K) (n = 6). ***P < 0.001 versus Pfkfb3WT_RA; ###P < 0.001 versus Pfkfb3WT_OIR. (L) Schematic illustration of the generation of myeloid and EC-specific Pfkfb3 double-KO (Pfkfb3ΔMϕΔEC) mice. (M) Representative retinal whole mounts from P17-OIR retinas of the indicated genotypes stained with isolectin B4 (red) with areas of NV highlighted (white). Two selected retinal areas (yellow box) were enlarged to show pathological neovessels. Quantification of pathological NV (N) in OIR retinas was expressed as percentage of total retinal areas. n = 10 to 13; Scale bars, 1000 μm for original images and 200 μm for enlarged images. ***P < 0.001. Data are means ± SEM. P value by Student’s t test [for (C), (H), and (I)] and one-way ANOVA followed by Bonferroni test [for (A), (B), (D), (F), (G), (K), and (N)].