Figure 1.
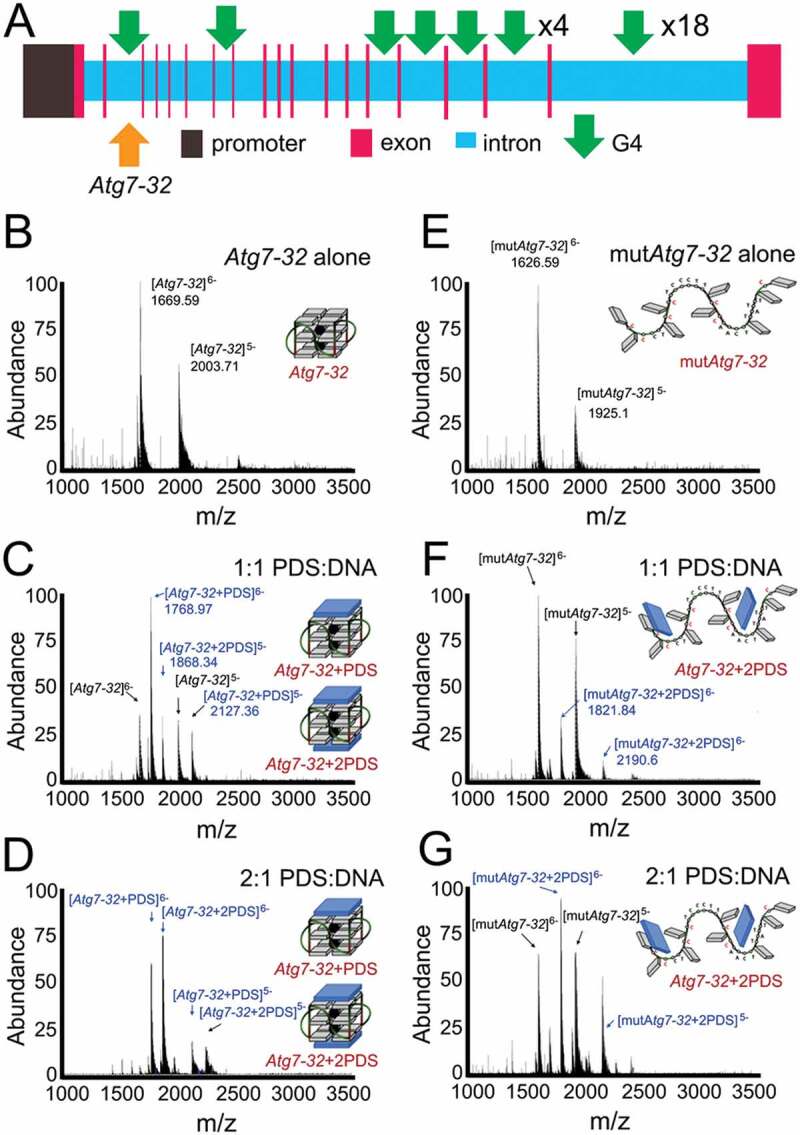
The stoichiometry and equilibrium binding constant of the non-covalent Atg7-32/PDS complexes. (A) A scheme of the rat Atg7 gene and its promoter showing putative G4-DNA locations. PQFSes in Atg7 and in its promoter were analyzed by using the QGRS mapper (http://bioinformatics.ramapo.edu/QGRS/index.php). 5,000 nt upstream the start codon were analyzed. (B-D) Electrospray ionization mass spectrometry (ESI-MS) experiments performed with Atg7-32 alone (10 µM) (B) or in presence of 1 and 2 mol. equiv. PDS (C and D, respectively). Mixtures were prepared in 100 mM ammonium acetate buffer and equilibrated at 25°C for 1 h prior to the experiments (20% of methanol added to the solution for the injection, performed at a flow rate of 10 μL/min). K = 4.82x105 M−1 (C), K = 1.47x107 M−1 (D). (E-G) ESI-MS experiments performed with mutAtg7-32 alone (10 µM) (E) or in presence of 1 and 2 mol. equiv. PDS (F and G, respectively). Mixtures were prepared in 100 mM ammonium acetate buffer and equilibrated at 25°C for 1 h prior to the experiments (20% of methanol added to the solution for the injection, performed at a flow rate of 10 µL/min). K = 2.19x104 M−1 (F), K = 7.73x104 M−1 (G)
