Figure 3.
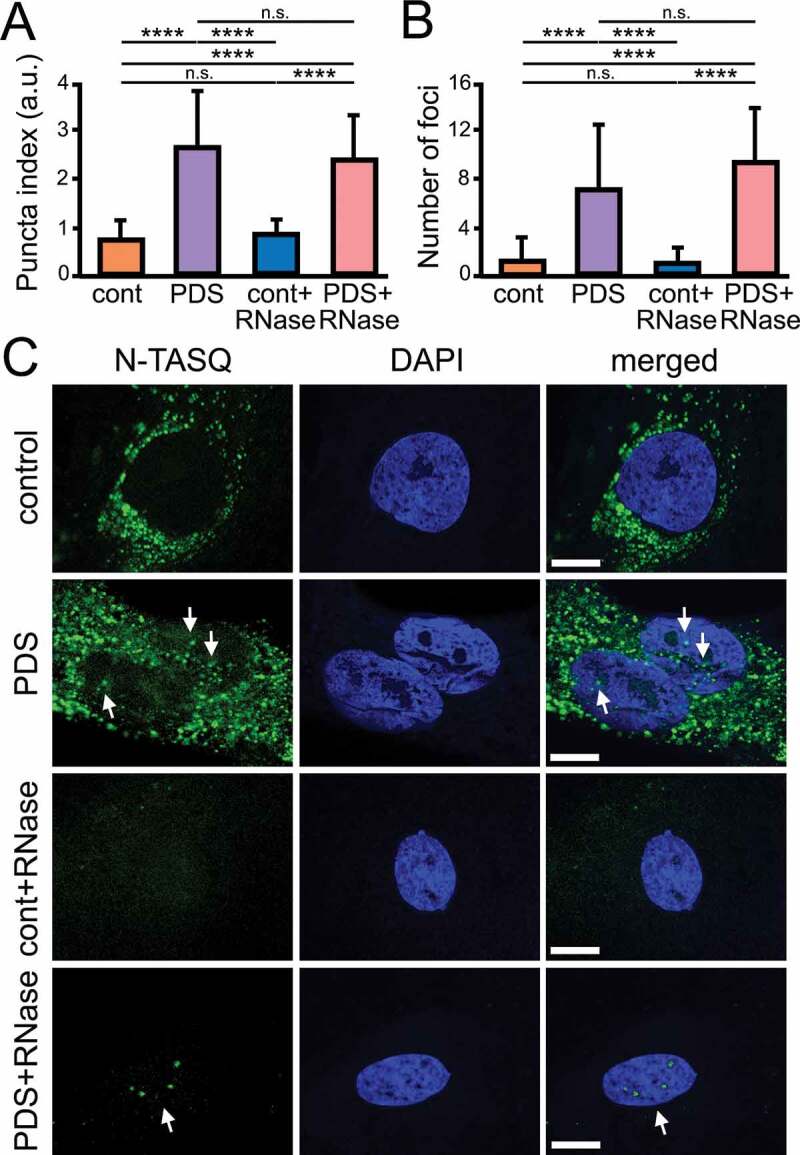
PDS alters G4 landscapes in astrocytes. (A) Cultured primary astrocytes were treated with a vehicle (control) or with PDS (2 μM) overnight. Cells were fixed and stained with N-TASQ (50 µM) and with the nuclear dye Hoechst (DAPI). Some samples were treated with RNase to digest RNA before staining. Fixed cells were automatically imaged with the EVOS microscope (N-TASQ and DAPI) and the nuclear puncta index was analyzed in all conditions. Two-way ANOVA followed by Tukey’s multiple comparisons test were used. ****p < 0.0001, n.s., non-significant. For each experiment, 80 cells were blindly analyzed, and results were pooled from two independent experiments. (B) The number of puncta was analyzed in cells from (A) with ImageJ. Two-way ANOVA followed by Tukey’s multiple comparisons test were used. ****p < 0.0001, n.s., non-significant. (C) Cultured primary astrocytes were treated with a vehicle (control) or with PDS (2 μM) overnight. Cells were fixed and stained with N-TASQ (50 µM) and with the nuclear dye Hoechst (DAPI). Some samples were treated with RNase to digest RNA before staining. Fixed cells were analyzed with a confocal microscope (N-TASQ and DAPI). Scale bar: 1 µm
