Figure 11.
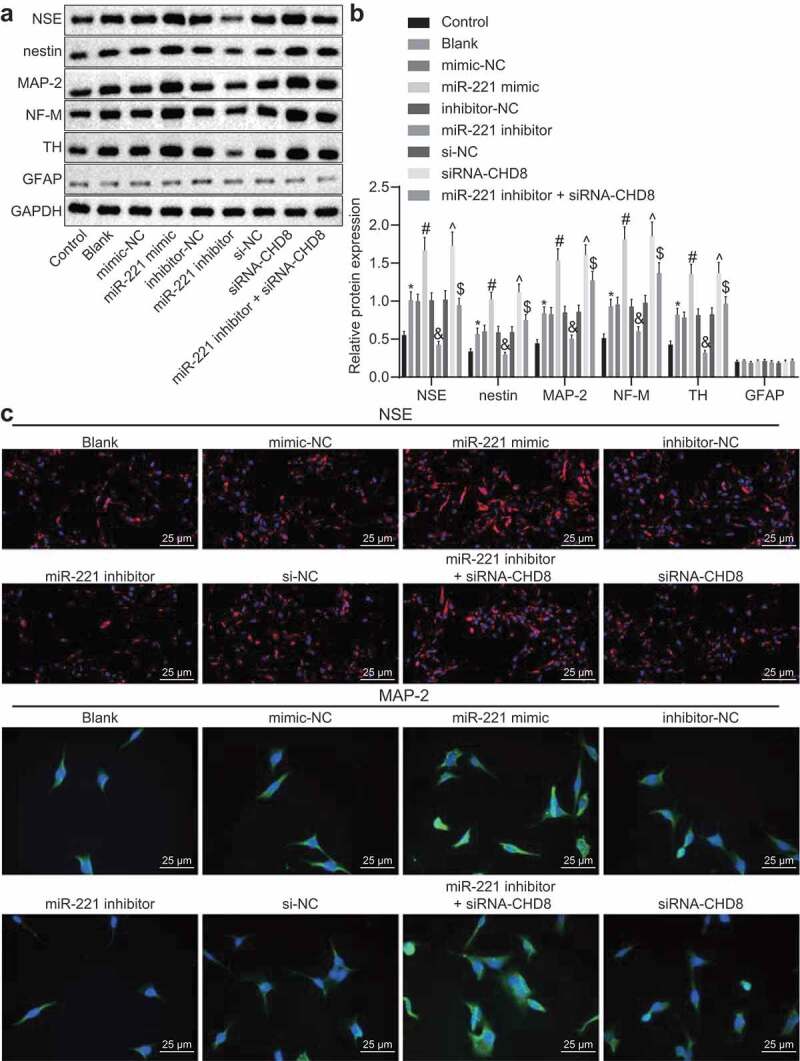
miR-221 accelerated the differentiation of SHEDs to neurons through suppression of CHD8. (a and b) The expressions of NSE, nestin, MAP-2, NF-M, TH, and GFAP in each group. determined by Western blot analysis. (c) The expressions of NSE (red) and MAP-2 (green) examined by immunofluorescence (×400). *, p < 0.05 vs. the control group; #, p < 0.05 vs. the mimic-NC group; &, p < 0.05 vs. the inhibitor-NC group; ^, p < 0.05 vs. the si-NC group; $, p < 0.05 vs. the miR-221 inhibitor group. Measurement data were expressed as mean ± standard deviation. One-way analysis of variance (ANOVA) was adopted to analyze comparison between multiple groups with Tukey’s post hoc test. The experiment was repeated three times. miR-221, microRNA-221; SHEDs, Stem cells from Human Exfoliated Deciduous teeth; CHD8, Chromodomain-Helicase-DNA-binding protein 8; NSE, Neuron Specific Enolase; MAP-2, Microtubule-associated protein-2; NF-M, midsized neurofilament; TH, tyrosine hydroxylase; GFAP, Glial fibrillary acidic protein; NC, negative control
