Figure 5.
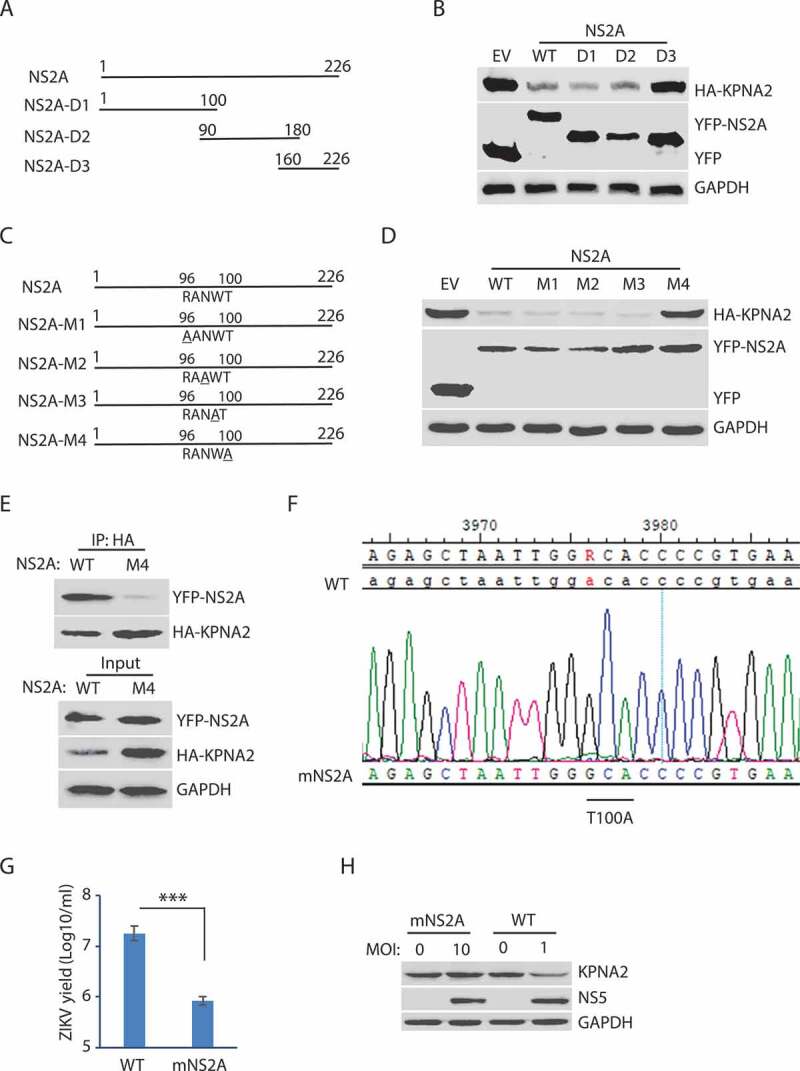
The amino acid residue T100 of NS2A is indispensable for the NS2A-mediated reduction of KPNA2. (A) Schematic illustration of the NS2A truncation constructs. The numbers above the lines indicate NS2A amino acid residues. (B) WB of KPNA2 in HEK293 cells co-transfected with plasmids of NS2A truncation constructs and HA-KPNA2. Wild-type (WT) NS2A and EV were included as controls. (C) Schematic illustration of mutant NS2A with point mutations. The amino acid residues of 96–100 are shown below the top line. Underlined residues indicate point mutations. M1: R96A, M2: N98A, M3: W99A, and M4: T100A. (D) WB of KPNA2 in HEK293 cells co-transfected with plasmids of mutant NS2A with point mutations and HA-KPNA2. WT NS2A and EV were included as controls. (E) Mutant NS2A-M4 (T100A) has minimal interaction with KPNA2. HEK293 cells were co-transfected with plasmids of HA-KPNA2 and WT NS2A or NS2A-M4. The input of cell lysate was included in WB. (F) Sequencing of the cDNA of the mutant ZIKV with NS2AT100A confirms the presence of codon mutation in its genome. The numbers above the sequence indicate nucleotide positions in cDNA of the ZIKV genome. The bar below the chromatogram indicates the codon change of T100A in NS2A. (G) Mutant ZIKV ICD (mNS2A) has a much lower proliferation than the WT virus. Vero cells were inoculated with an MOI of 0.01 for ZIKV proliferation and harvested for virus titration at 72 hpi. A significant difference is denoted by *** for P< 0.001. (H) Mutant ZIKV has minimal effect on the KPNA2 level. Vero cells were inoculated with an MOI of 10 for the mutant mNS2A virus or with an MOI of 1 for the WT virus to ensure both mutant and WT viruses had a similar level of replication and harvested at 48 hpi for immunoblotting
