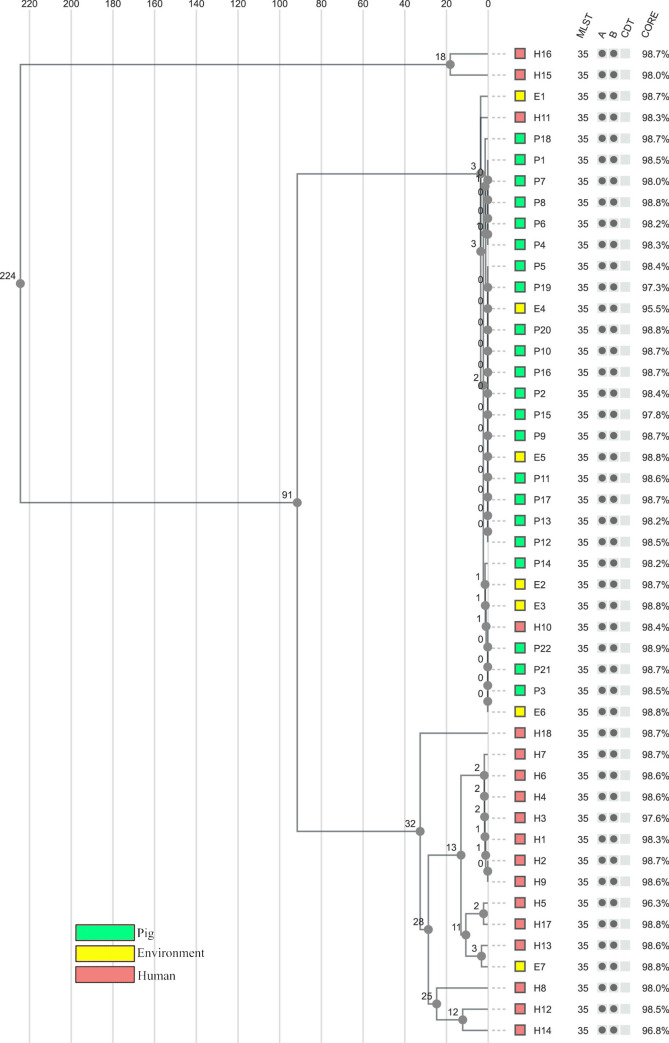
Fig 2. 1928D cgMLST analysis tree.
Unweighted pair group method with arithmetic mean (UPGMA) tree including all isolates with distances based on the 1928D core genome multilocus sequencing typing (cgMLST) scheme (2,631 genes), ignoring missing alleles. Coloured according to the isolation source. Sequence type (MLST), the presence of toxin genes (A = tdcA, B = tdcB, CDT = binary toxin), and percentage of good cgMLST genes (CORE) are presented after each isolate.