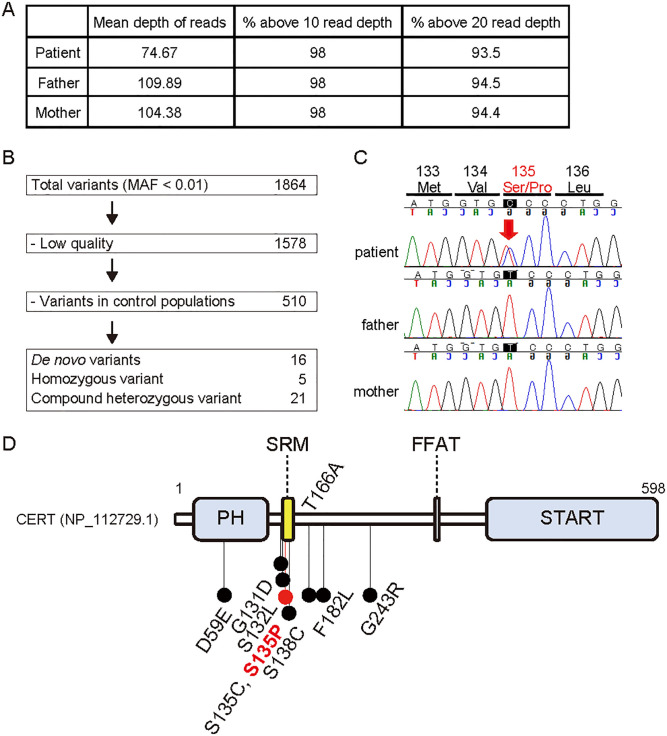
Fig 2. An ID-associated de novo CERT1 mutation.
(A) WES sequencing quality. (B) Filtering pipeline of variants identified by trio WES. A total of 1864 variants with a minor allele frequency (MAF) of less than 0.01 were found in the patient. After excluding low-quality variants and variants found in the control healthy population, 16, 5, and 21 variants were identified as de novo heterozygous, homozygous, and compound heterozygous, respectively. (C) Trio Sanger sequence of the CERT1 variant (NM_0031361.3:c.403T>C:p.[Ser135Pro], Chr5[GRCh37]:g.74722249A>G) identified in our patient. (D) Variant distribution of the CERT protein structure (NP_112729.1), which includes the PH domain, SRM, FFAT motif, and steroidogenic acute regulatory protein-related lipid transfer (START) domain.