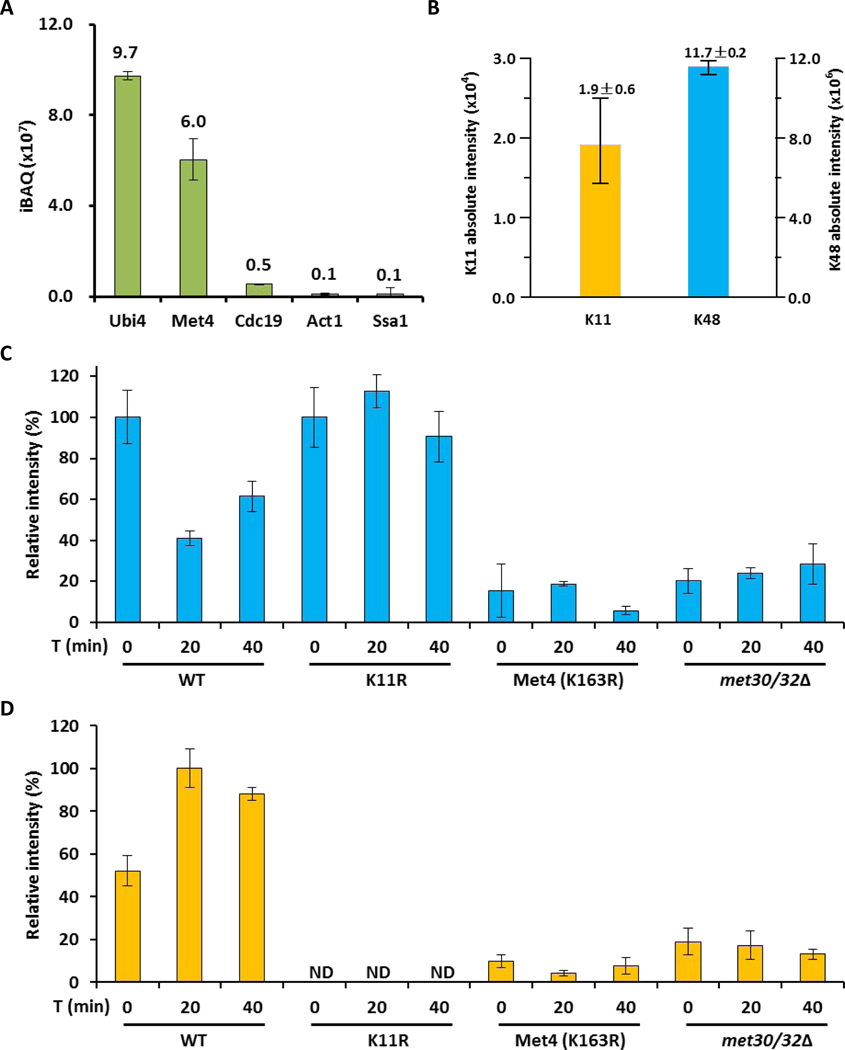
Figure 4. The topology of the Met4 attached ubiquitin chain changes from K48 to K11 during activation.
(A) Intensity distribution of co-IP purified proteins with 3HA-tagged Met4 proteins.
(B) Ubiquitin linkages on 3HA-tagged Met4 proteins identified by LC–MS.
(C) K48-linked chains on 3HA-tagged Met4 proteins during metabolic shift to methionine depleted medium were quantified by LC–MS. The amount was normalized based on the sample from methionine containing medium. Results are shown for mean +/− SEM. Samples were prepared from wild-type cells, K11R mutants, Met4K163R, and met30Δ met32Δ double mutants expressing cells. The met32Δ deletion was required to suppress the lethality of met30Δ mutants.
(D) Same as panel C, but the K11-linkage was quantified.