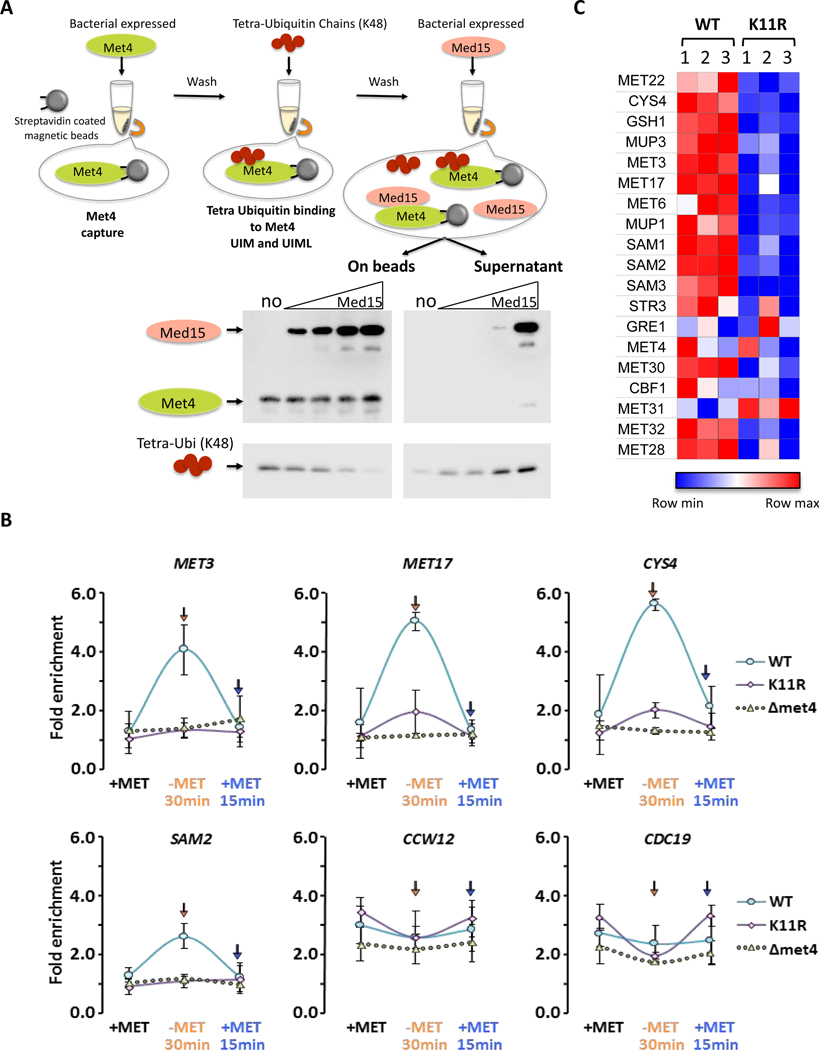
Figure 6. K48 and K11 linked ubiquitin chains on Met4 control expression through modulation of mediator binding.
(A) K48 ubiquitin chains and the mediator component Med15 have overlapping, exclusive binding sites. Immobilized Met4 (76–160) was bound to K48 tetraubiquitin. Elution of K48 tetraubiquitin by addition of 0, 0.25, 0.5, 1, or 3-fold molar excess of Med15 (residues Med15 1–651 Δ239–271, Δ373–483) was analyzed by immuno blotting of the eluted fraction and the bead bound fraction.
(B) Mediator recruitment to MET gene promoters depends on K11 ubiquitin chains. Wild-type, K11R, and Δmet4 mutants were cultured in medium containing repressive amounts of methionine, shifted to methionine depleted growth medium for 30 minutes to activate MET gene transcription before re-repression by addition of methionine for 15 minutes. Mediator binding was monitored by ChIP using the endogenously HA3-tagged mediator subunit Med14. Four Met4-dependent genes (MET3, MET17, CYS4, SAM2) and two Met4-independent genes (CCW12, CDC19) were analyzed. Results are shown for n=3 +/− SD.
(C) Yeast cells lacking the ability to form K11-linked ubiquitin chains (K11R mutants) show a significant defect in activation of Met4 controlled genes involved in methionine biosynthesis. RNA abundance of a panel of MET genes was analyzed on a custom NanoString panel. Three biological replicates are shown. Data were normalized to three housekeeping genes.