Fig. 4 |. Two-step alignment method.
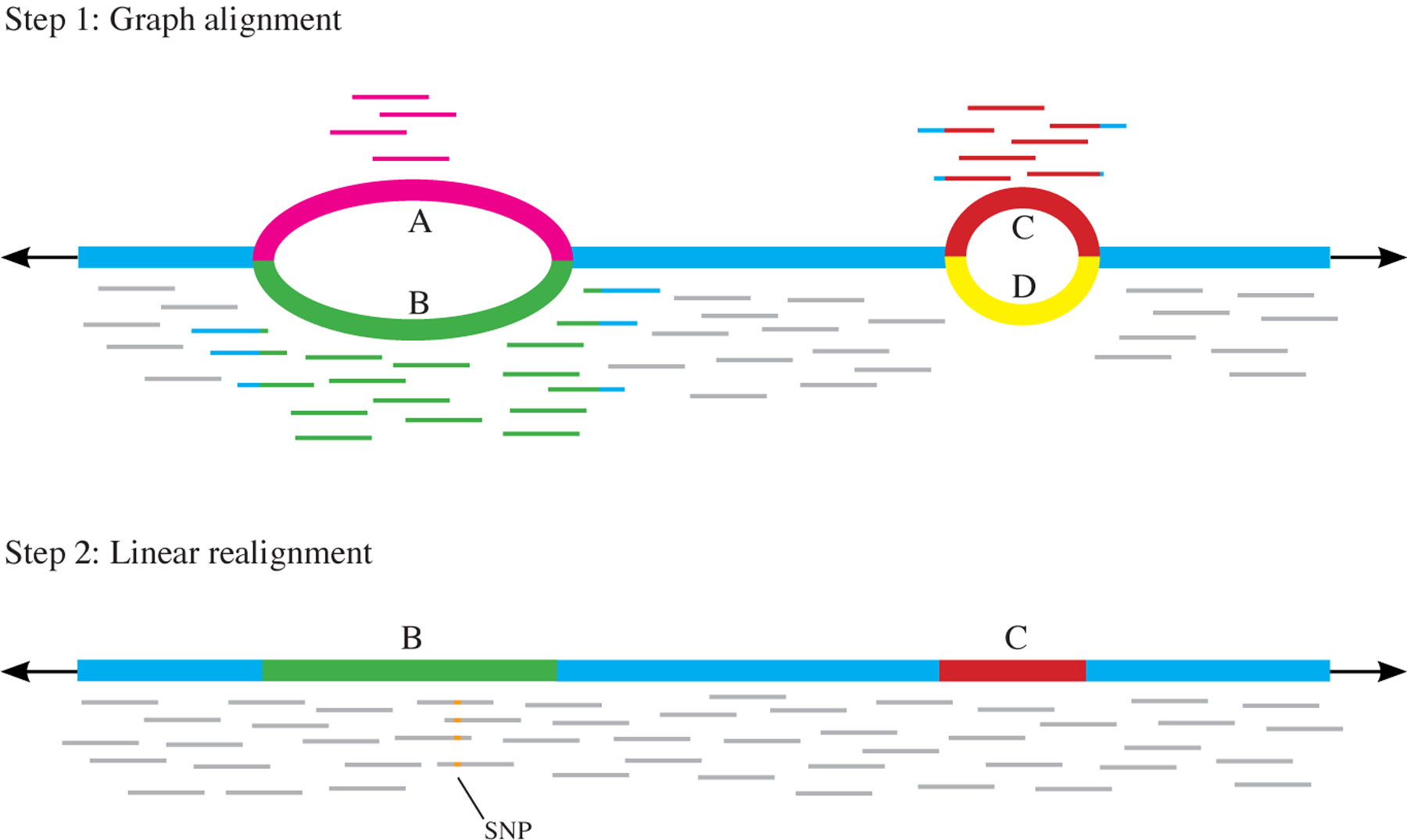
First, alignment to a graph is performed. Reads can align to either variant A or B at the first variant locus, and to C or D at the second. The path through the graph with the most reads aligned to it is then extracted — in this case, the path containing B and then C. In the second step, reads are realigned to the extracted linear genome. This allows for reads that may have been misaligned in the initial step (due to the introduction of variants) to be realigned only to the alleles they are most likely to have originated from. Here, the four reads that aligned to variant A now align to variant B, allowing a single-nucleotide polymorphism (SNP) to be detected that was undetectable from the graph alignment alone, as the reads with the SNP were misaligned.
