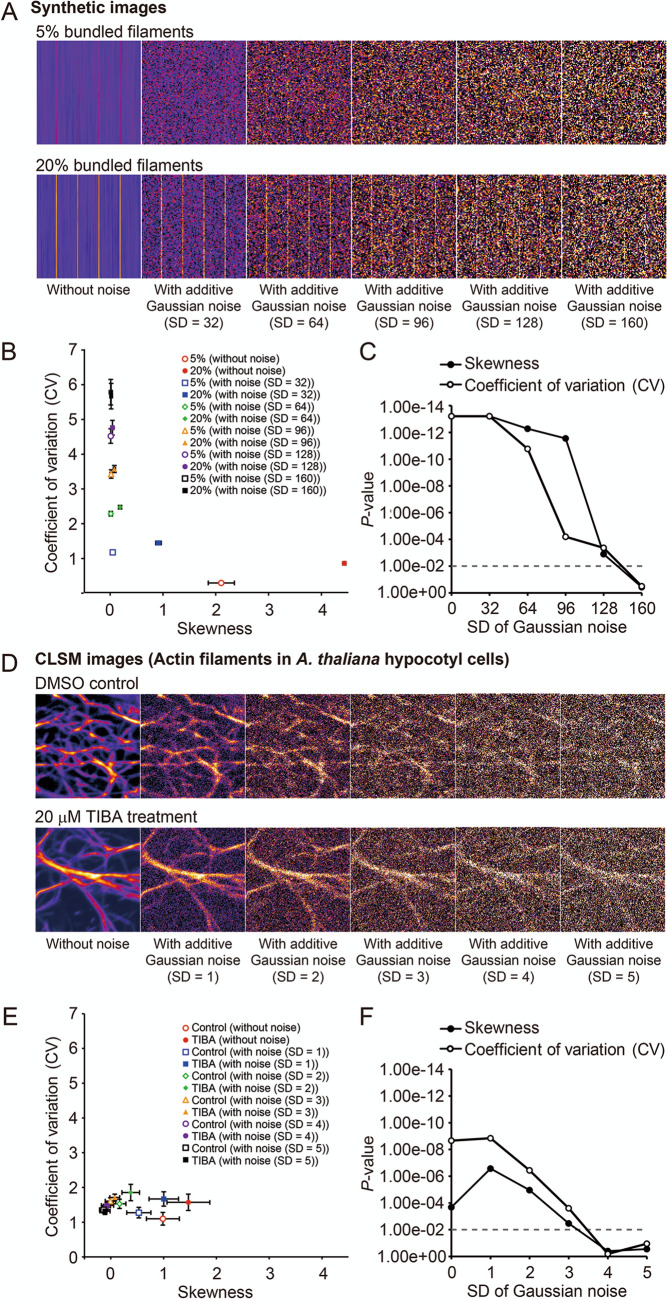
Figure 9.
Effects of noise on evaluations of cytoskeleton bundling in synthetic images and actual CLSM images. (A) Original and synthetic images of virtual cytoskeleton bundling with random Gaussian noise. Random Gaussian noise [average intensity = 28, standard deviation (SD) = 32–160] was added to the images with 5% and 20% bundled filaments. (B) Scatter plot between skewness and CV. (C) Scatter plot showing the sensitivity of the skewness and CV methods in the detection of bundles in synthetic images with added noise. P values between the images with 5% and 20% bundled filaments were determined using the Mann–Whitney U-test. The broken line indicates the significance level (P = 0.01). (D) Original and confocal laser scanning microscopy (CLSM) images of GFP-ABD2-labeled actin filaments in hypocotyl cells of A. thaliana plants. The image intensity was first normalized (average intensity = 0, standard deviation (SD) = 1), then Gaussian noise (average intensity = 0, SD = 1–5) was added to the actin filaments treated with DMSO (control images) and 20 μM TIBA (treatment images). Before measuring the skewness and CV, the images were skeletonized. (E) Scatter plot between skewness and CV. (F) Scatter plot showing the sensitivity of the skewness and CV methods in the detection of bundles in the CLSM images with added noise. P values were determined using the Mann–Whitney U-test. The broken line indicates the significance level (P = 0.01).