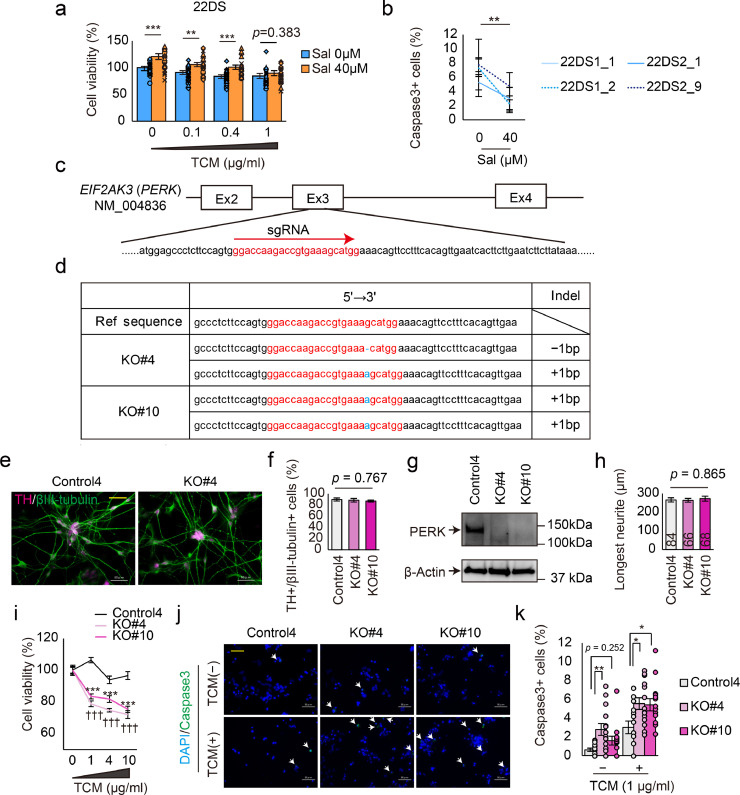
Fig. 4.
Generation of PERK-deficient iPSCs. (a) Cell viability in the presence of TCM with or without salubrinal (Sal). n = 24 (22DS1_1, n = 6; 22DS1_2, n = 6; 22DS2_1, n = 6; 22DS2_9, n = 6). Each plot represents the value of each line. The bars represent means ± SEs. **p < 0.01, ***p < 0.001. (b) Quantification of the caspase 3-positive (+) cell ratio using the 22DS group. TCM treatment (0.4 μg/ml) with or without Sal. Twenty fields were used (each, n = 5). The plots represent means ± SEs. ** p < 0.01. (c) The target site of the CRISPR/Cas9 system used in this study. (d) Indel patterns of the PERK-deficient isogenic lines. (e) Representative images of dopaminergic neurons (Day 24) immunostained for TH and βIII-tubulin. The scale bar represents 50 μm. (f) Analysis of dopaminergic neuron differentiation efficiency via the quantification of the ratio of TH+ to βIII-tubulin+ cells at Day 24. The bars represent means ± SEs. Five fields were used. (g) Immunoblotting of PERK and β-actin using DA neurons at Day 24. (h) Measurement of neurite length. The numbers indicated in the bars represent the number of counted cells. The bars represent means ± SEs. (i) Cell viability in the presence of ER stress (n = 7). The plots represent the means ± SEs. *** p < 0.001, ††† p < 0.001 vs. without TCM. (j) Representative images of immunostaining for cleaved-Caspase3 using dopaminergic neurons. White arrows indicate Caspase3+ cells. The yellow scale bar in the image represents 50 μm. (k) Quantification of the cleaved-caspase 3+ cell ratio (n = 14). The bars represent means ± SEs. * p < 0.05, ** p < 0.01. Each plot represents the value of each line.