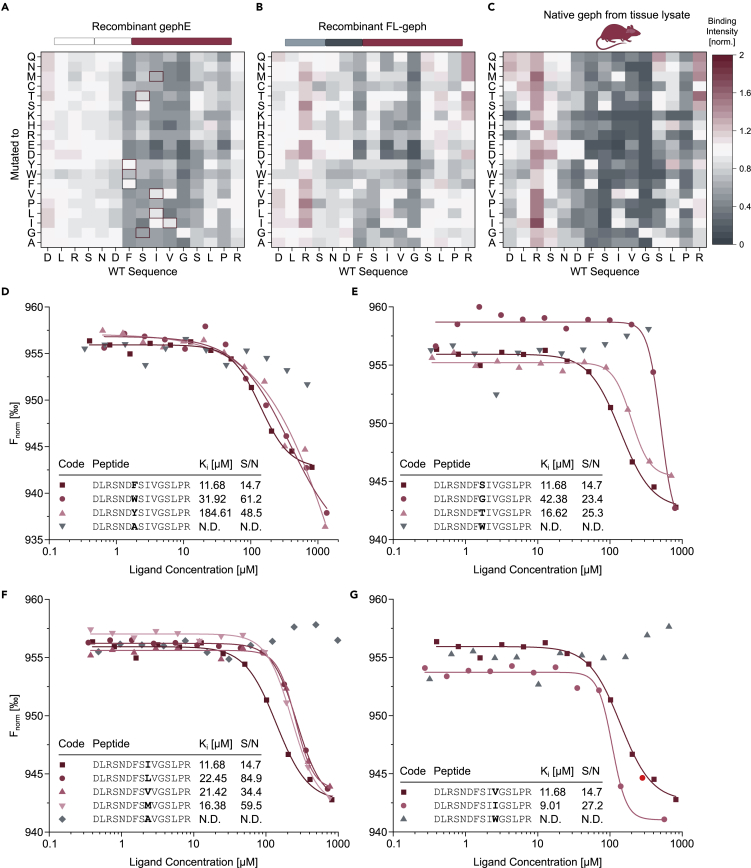
Figure 5.
Comparison between binding profiles of GlyR β to gephE, FL-geph, and native Geph in μSPOT and TRIC format
(A) A full positional scanning library was probed with recombinant gephE, FL-geph (B), and native geph from tissue lysate (C) in microarray format. Geph domain architectures of the applied recombinant protein or lysate origin are indicated above the heatmaps. Shown are intensity values of point-mutated variants normalized to the corresponding WT sequence (GlyR β 414DLRSNDFSIVGSLPR428) displayed as a heatmap. Higher spot intensity corresponds to preferential binding, vice versa. In A, brackets corresponding to peptides that exhibited a determinable Ki value in TRIC format are bordered in red, whereas a representative WT sequence is bordered in blue. Values are presented as mean of n = 3. Refer to Table S5 for STDEV of each bracket.
(D–G) Determined dose responses and derived Ki values of peptide variants in TRIC format. The interchanged position (position 420 (D), 421 (E), 422 (F) and 423 (G) of the GlyR β subunit respectively) is highlighted in bold. Peptides with a quantifiable Ki value are shown in red, alongside a representative non-binding variant in gray. Corresponding brackets in (A) are bordered in red. Bright red dots were excluded from curve fitting. Refer to Table S6 for starting concentrations of each peptide and Figure S7 for a direct comparison of the binding intensity found in microarray format and Ki values determined using TRIC.