FIGURE 1.
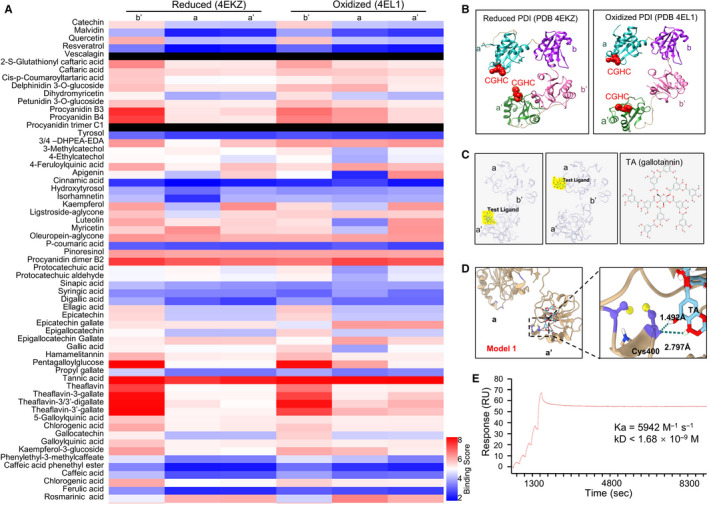
Tannic acid binds PDI molecule with high affinity. SystemsDock website server was used to simulate the interaction between TA and PDI molecular at different oxidative status. A, Heat map of 61 plant‐derived polyphenolic compounds that potentially bind to different domains (b′, a and a′) of reduced (left three columns) or oxidized (right three columns) PDI molecules. Colour bars reflects the rank of binding scores. B, The protein structure of human PDI at reduced and oxidized status was obtained from the PDB database. Red spheres, CGHC active centre. C, Molecular docking results of TA (yellow) binding to the a′ (left panel) and a (middle panel) domain of PDI molecule (grey) from systemsDock and two‐dimensional molecular structure of TA (right panel). Red dot in TA structure, hydroxyl groups. D, Molecular simulation result of TA binding with the a′ domain active centre. Purple structures, the cysteine residues Cys397 and Cys400 at the CGHC active site in the a′ domain of PDI. Yellow sphere, hydroxyl group. The green dotted line in the right panel, hydrogen bond formed between Cys400 and TA. E, Binding of soluble TA in flow with immobilized PDI on biochip was quantified using the surface plasmon resonance assay. SPR sensorgram (red line) showed a strong binding between TA and PDI
