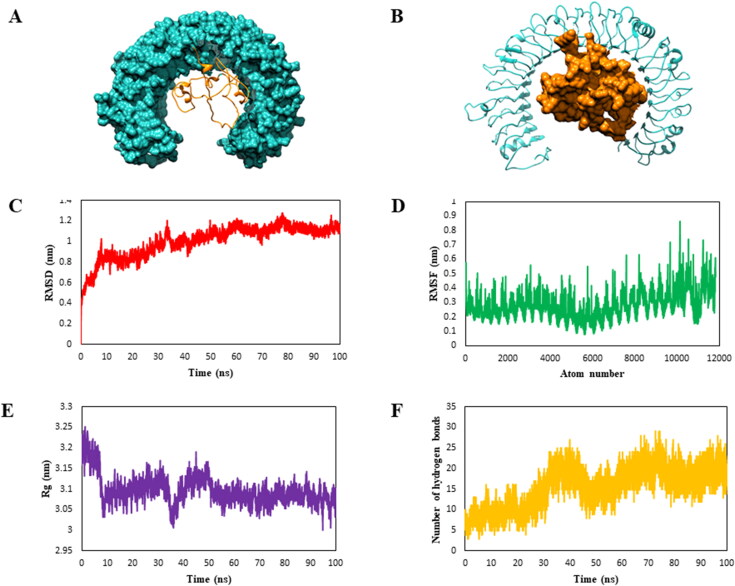
Figure 5.
Molecular docking and molecular dynamics simulations studies. (A and B) The molecular interaction studies of vaccine candidate with immune receptor TLR3 using HDOCK server was performed and different representation of the vaccine (orange) and TLR3 receptor (orange) are shown here. Molecular dynamics simulation of vaccine and immune receptor TLR3 complex was done for 100 ns using GROMACS. (A) RMSD (root mean square deviation) plots reflect the stability between the vaccine candidate and TLR-3 receptor whereas (B) RMSF (root mean square fluctuation) reflecting the flexibility and fluctuation of the residual atoms in the side chain of docked complex. (C) Radius of gyration is plotted as a function of time to know the structural compactness. (D) Hydrogen bonds of the complex are graphically represented to study the changes in stability of vaccine-receptor complex.