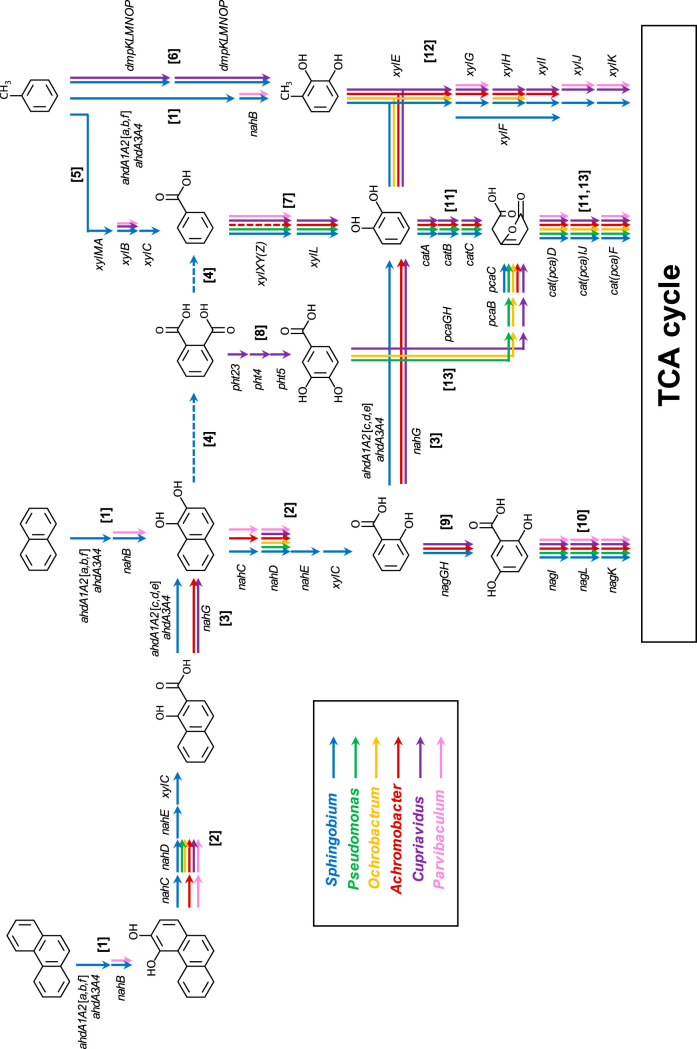
FIG 5.
Distributions of key aromatic hydrocarbon degradation genes harbored in six high-quality MAGs for Sphingobium, Pseudomonas, Ochrobactrum, Achromobacter, Cupriavidus, and Parvibaculum spp. reconstructed from the consortium grown on diesel fuel. Arrows labeled as different colors indicate functional genes found in each MAG. (Step 1) Initial biotransformation step of phenanthrene, naphthalene, and toluene by class A ARHDs. (Step 2) Transformation of 3,4-dihydroxyphenanthrene and 1,2-dihydroxynaphthalene via extradiol ring cleavage pathways. (Step 3) Decarboxylation of 1-hydroxy-2-naphthoic acid and salicylic acid. (Step 4) Transformation of 1,2-dihydroxynaphthalene via intradiol ring cleavage pathway. (Step 5) Methyl-monooxygenation of toluene. (Step 6) Ring-monooxygenation of toluene. (Step 7) Transformation of benzoic acid to catechol. (Step 8) Transformation of phthalic acid to protocatechuic acid. (Step 9) Transformation of salicylic acid to gentisic acid. (Step 10) Degradation of gentisic acid. (Steps 11 and 12) Degradation of catechol via intradiol or extradiol ring cleavage pathways. (Step 13) Degradation of protocatechuic acid.