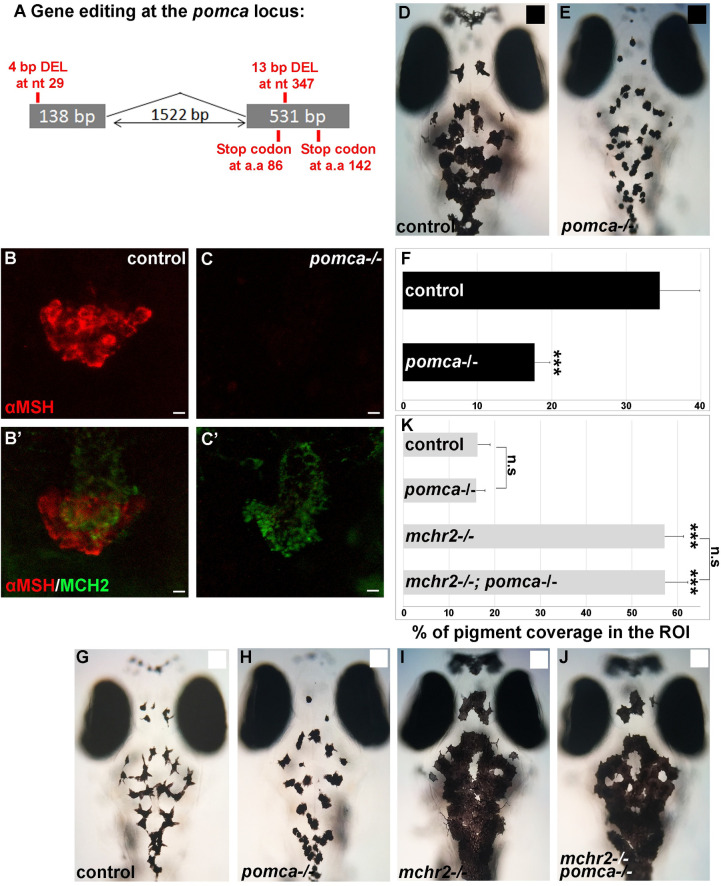
Fig 5. pomca loss of function leads to melanosomes contraction, but is not sufficient to rescue the mchr2 mutant phenotype in larvae.
(A) Schematic representation of the pomca locus and the small genomic deletions induced by CRISPR/Cas-9 leading to premature stop codons in the pomca coding sequence. (B-C’) Confocal projection (B, C) or section (B’, C’) of double immunolabelling with MCH2 and α-MSH antibody at 5 dpf, showing that MCH expression, but not α-MSH, is detected in the pomca homozygous mutant. (D, E) Dorsal melanocytes of control (D) or pomca homozygous mutant (E) black adapted larvae at 7 dpf. (F) Melanosome coverage was quantified in control or pomca homozygous mutant black adapted larvae at 7 dpf. (G-J) Dorsal melanocytes of control (G), pomca homozygous mutant (H), mchr2 homozygous mutant (H) or pomca; mchr2 double homozygous mutant (J) white adapted larvae at 7 dpf. (K) Melanosome coverage was quantified in control, pomca homozygous mutant, mchr2 homozygous mutant or pomca; mchr2 double homozygous mutant white adapted larvae at 7 dpf. Dorsal view with anterior up (D, E and G-J). Scale bars: 10 μm. Error bars represent s.d. *P<0.05, **P<0.001, ***P<0.0005, determined by t-test, two-tailed.